1.4. Access to datasets in the database
When you access https://jmorp.megabank.tohoku.ac.jp, the top page of the jMorp website appears.
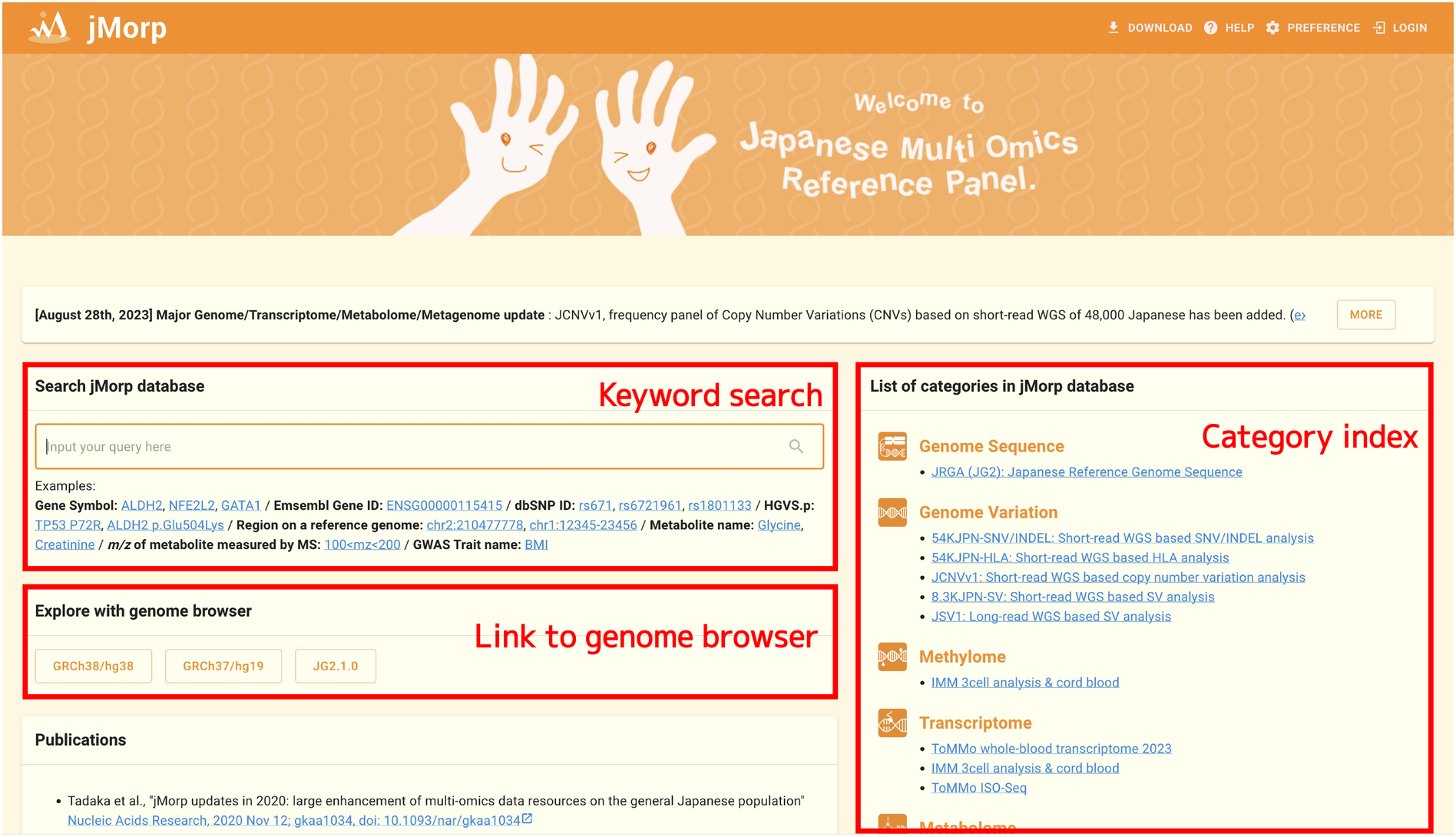
There are roughly three blocks on the top page (keyword search, genome browser, and category index).
1.4.1. Keyword search
You can use the keyword search panel in the upper left corner to conduct a keyword search on all of the data in the jMorp. You can use the following kinds of keywords:
Note
To navigate to the actual search results page, click on any of the example search keywords below. Please give it a try to see what kind of page you get.
Category |
Query example |
|---|---|
Gene Symbols |
|
Ensembl Gene ID |
|
UniProt ID |
|
PDB ID |
|
dbSNP ID (rsID) |
|
Gene symbol + HGVS.p, RefSeq ID + HGVS.p |
TP53 P72R / ALDH2 p.Glu504Lys / NP_000537 P72R / NP_000681 p.Glu504Lys |
Position / Region on a reference genome |
|
Metabolite name |
|
m/z and retention time of metabolites analyzed by MS |
|
GWAS trait name |
At the time of this writing, for example, a search for Creatinine yields the following results:
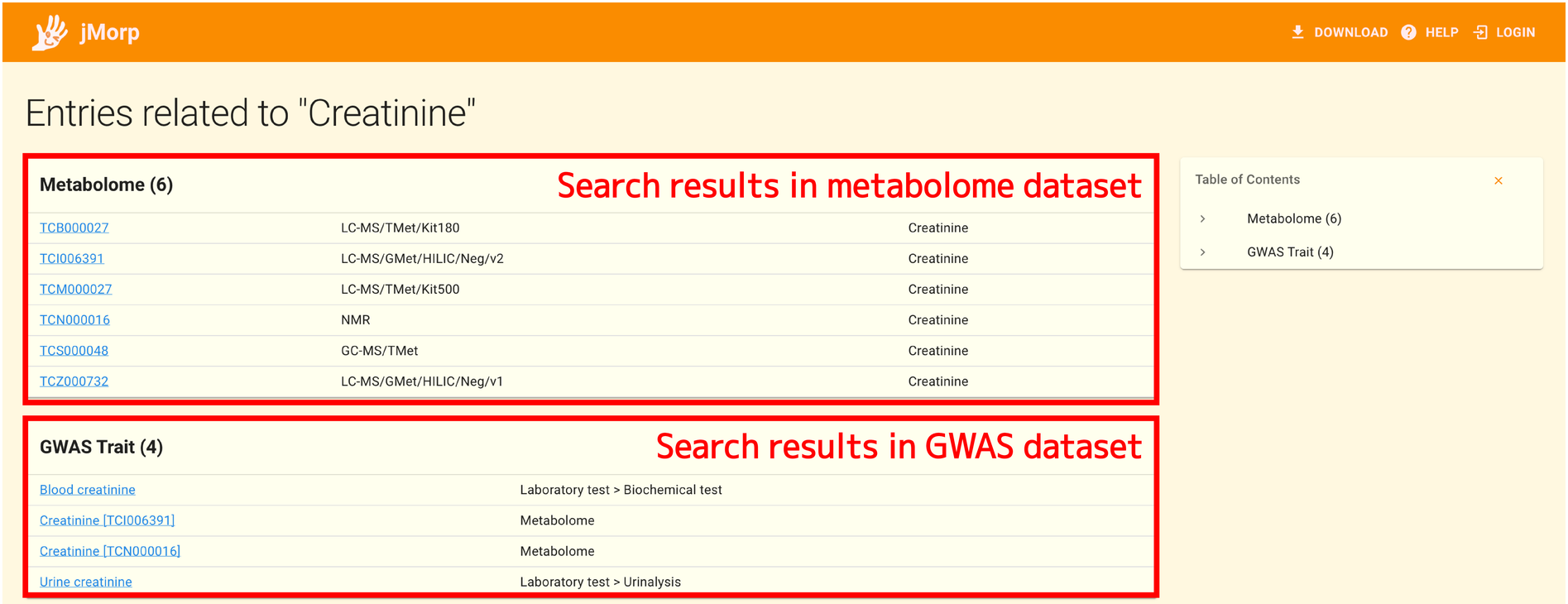
The search results from the metabolome dataset are displayed at the top of the search results page. In the metabolome dataset, creatinine was examined using a variety of techniques, including NMR, LC-MS, and GC-MS, and each of these analyses is represented as a separate entry.
The search results from the GWAS dataset are presented at the bottom of the search results page. You can view the results of analyses using Creatinine as a GWAS trait among the GWAS analysis results stored in jMorp by clicking the link in this panel. It is possible to conduct a cross-search on the data stored in jMorp, as in this example, by using the keyword search panel on the top page of jMorp.
Note
Since the keyword Creatinine matches multiple entries, the search result page like above is displayed. However, if an input keyword matches just one jMorp entry, the search result page is not shown, and you are instead taken directly to the entry page.
1.4.2. Genome browser view
You can browse the genome-related data in the genome browser by clicking the link on the bottom left of the jMorp top page. There are three different types of genome browsers available now: GRCh37/hg19, GRCh38/hg38, and JG2.1.0. The genome-related data stored in jMorp uses different reference genomes for analysis, and the data that can be browsed differs depending on the selection of genome browsers.
- GRCh37/hg19
- Genome Variation
3.5KJPNv2: SNV/INDEL frequency panel derived from short-read whole genome analysis of 3,500 samples
4.7KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 4,700 samples
8.3KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 8,300 samples
8.3KJPN-SV: Structual variation (SV) frequency panel derived from short-read whole genome analysis of 8,300 samples
- Genome (others)
Genome Accessibility: Mean depth data calculated from TMM short-read whole genome sequencing data
- GRCh38/hg38
- Genome Variation
14KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 14,000 samples
38KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 38,000 samples
54KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 54,000 samples
JCNVv1: CNV frequency panel derived from short-read whole genome analysis of 48,000 samples
JSV1: Structual variation (SV) frequency panel derived from long-read whole genome analysis of 222 samples
- Genome (others)
Genome Accessibility: Mean depth data calculated from TMM short-read whole genome sequencing data
- JG2.1.0
- Genome Sequence
JG2.1.0: JG2.1.0 Japanese reference genome sequence
- Genome Variation
38KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 38,000 samples (liftover from GRCh38 version)
54KJPN: SNV/INDEL frequency panel derived from short-read whole genome analysis of 54,000 samples (liftover from GRCh38 version)
For instructions on using the genome browser, see Genome browser.
1.4.3. Category index
There is a list of the datasets that are part of jMorp on the right side of the top page. To view the list of entries in a dataset, click on the dataset name. For example, clicking on Metabolome 2023 brings up the table below, which contains a list of the metabolites that are included in the metabolome dataset.
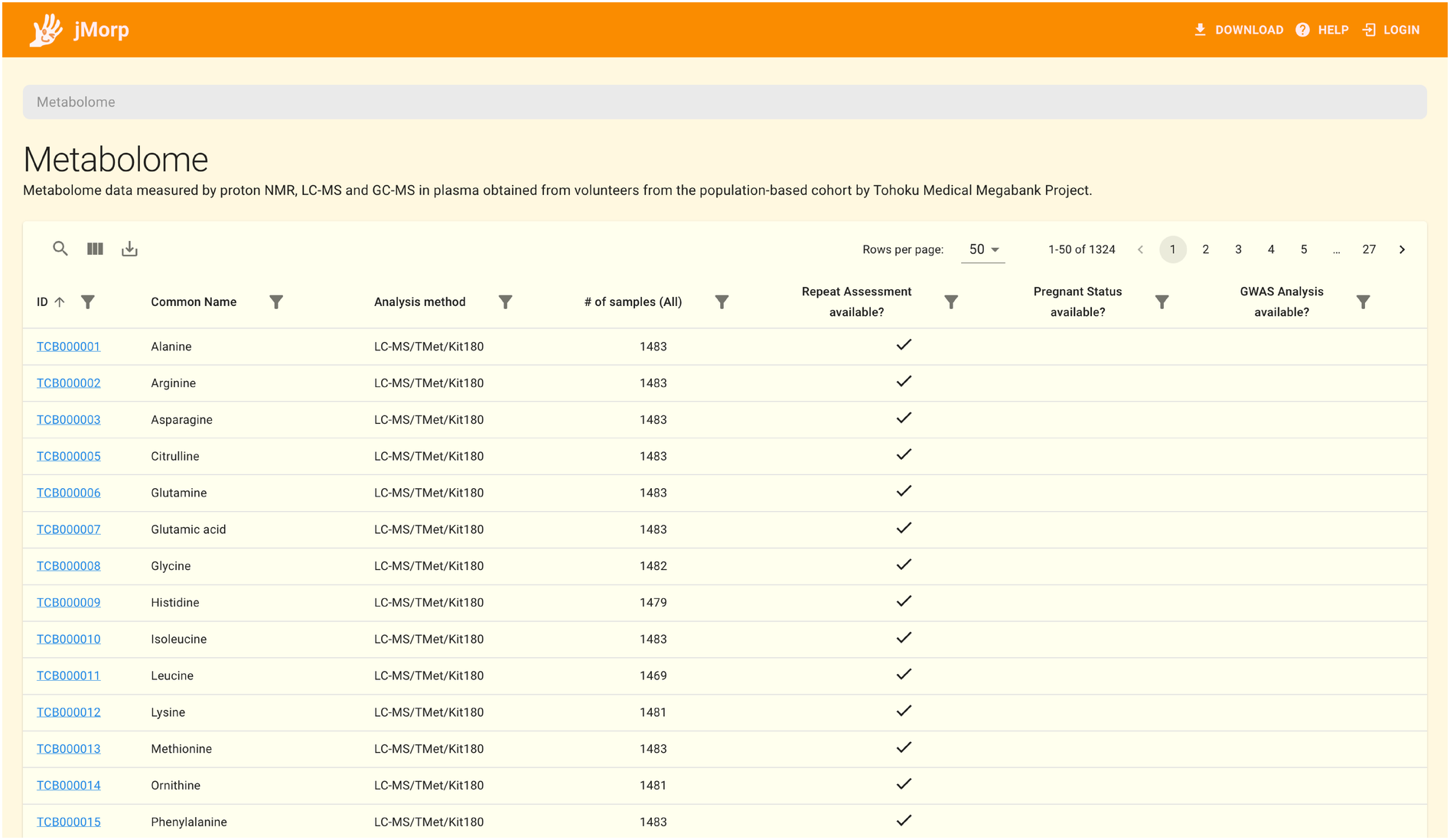
Additionally, you can browse the metabolite’s detailed information by clicking the metabolite ID.
The category index is the best option if you want to quickly see what kind of data is contained in jMorp.
Note
For categories with a lot of entries, like Genome Variation (SNV/INDEL), lists of entries are not available. Please make use of the keyword search and genome browser views.