2.7. Genome Browser
jMorp has a genome browser that allows you to browse the whole genome reference panel, Japonica array marker locations, and GWAS results while checking their positional relationships on the genome. This genome browser is built on JBrowse2.
2.7.1. Open Genome Browser
The genome browser can be opened from the “Explore with genome browser” panel on the top page. The three assemblies: “GRCh38/hg38”, “GRCh37/hg19”, and “JG2.1.0” are available.
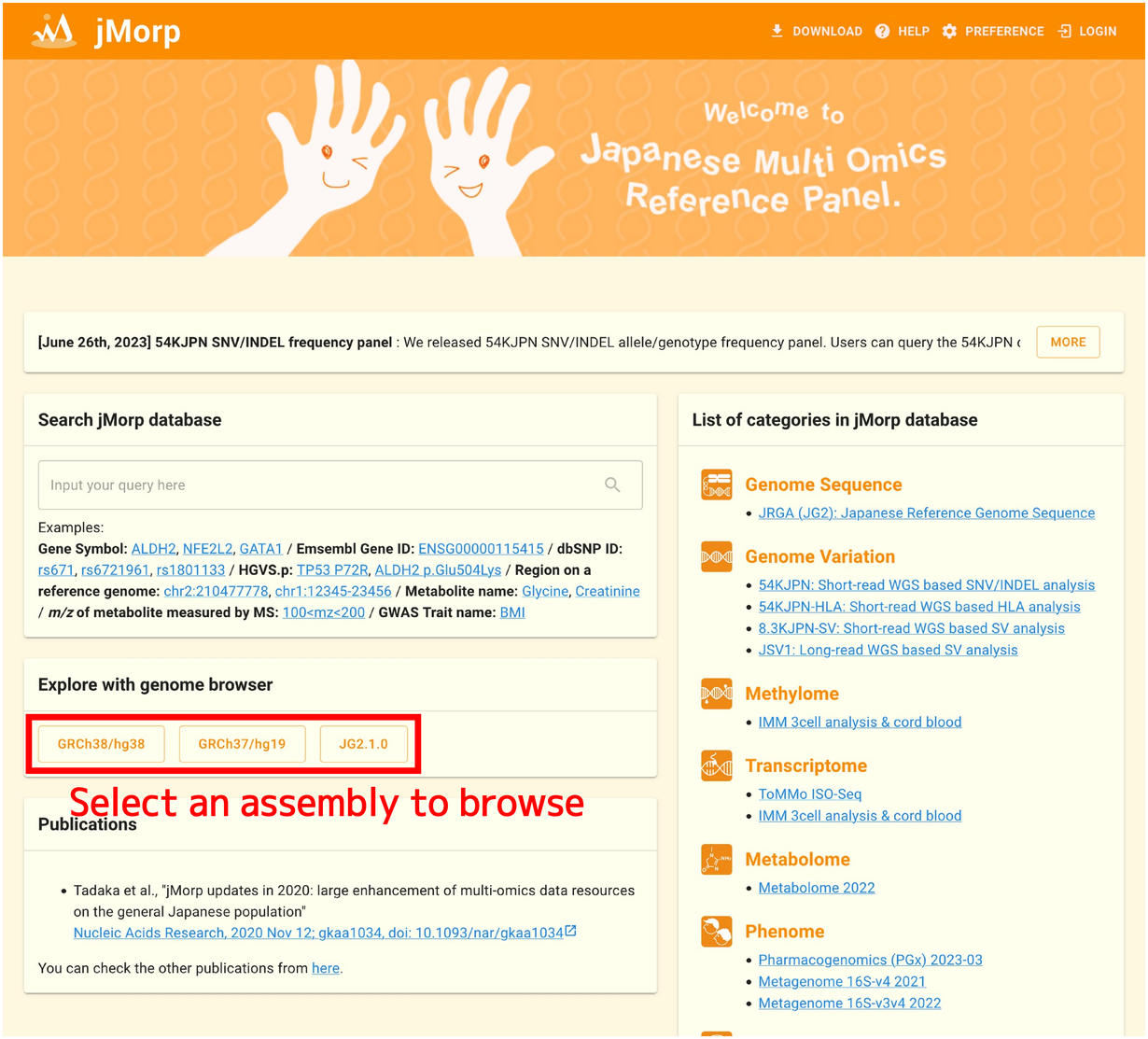
Each assembly provides different data. Examples are shown below.
Assembly |
Provided Data |
|---|---|
GRCh38/hg38 |
|
GRCh37/hg19 |
|
JG2.1.0 |
|
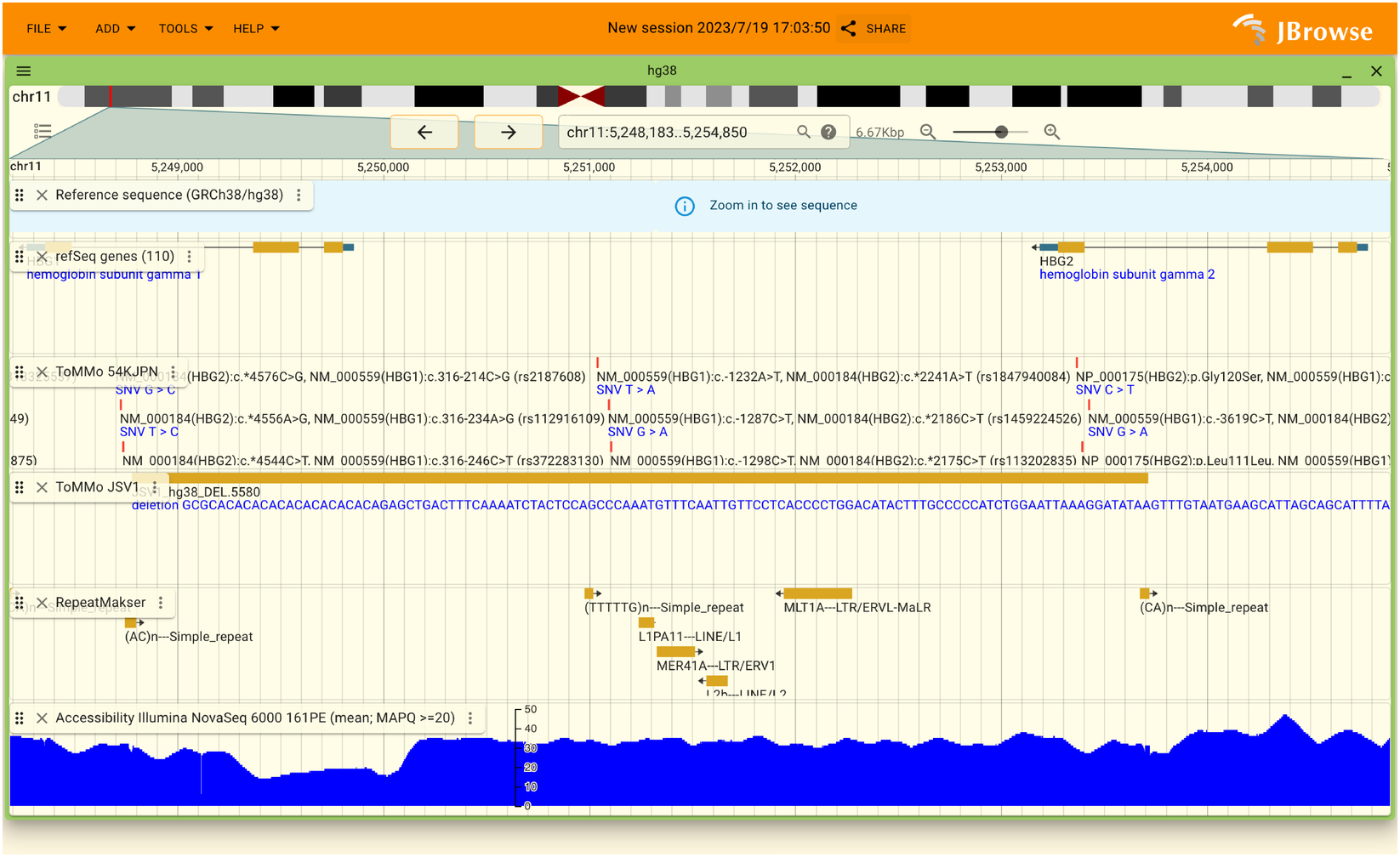
When the genome browser is successfully loaded, the following display will appear.
2.7.2. The list of tracks
When you opened the genome browser, some limited number of contents are shown. We provide a lot of data about TMM and other annotation and can be shown in the genome browser.
To display the list of tracks, please click a red rectangle shown in the below.
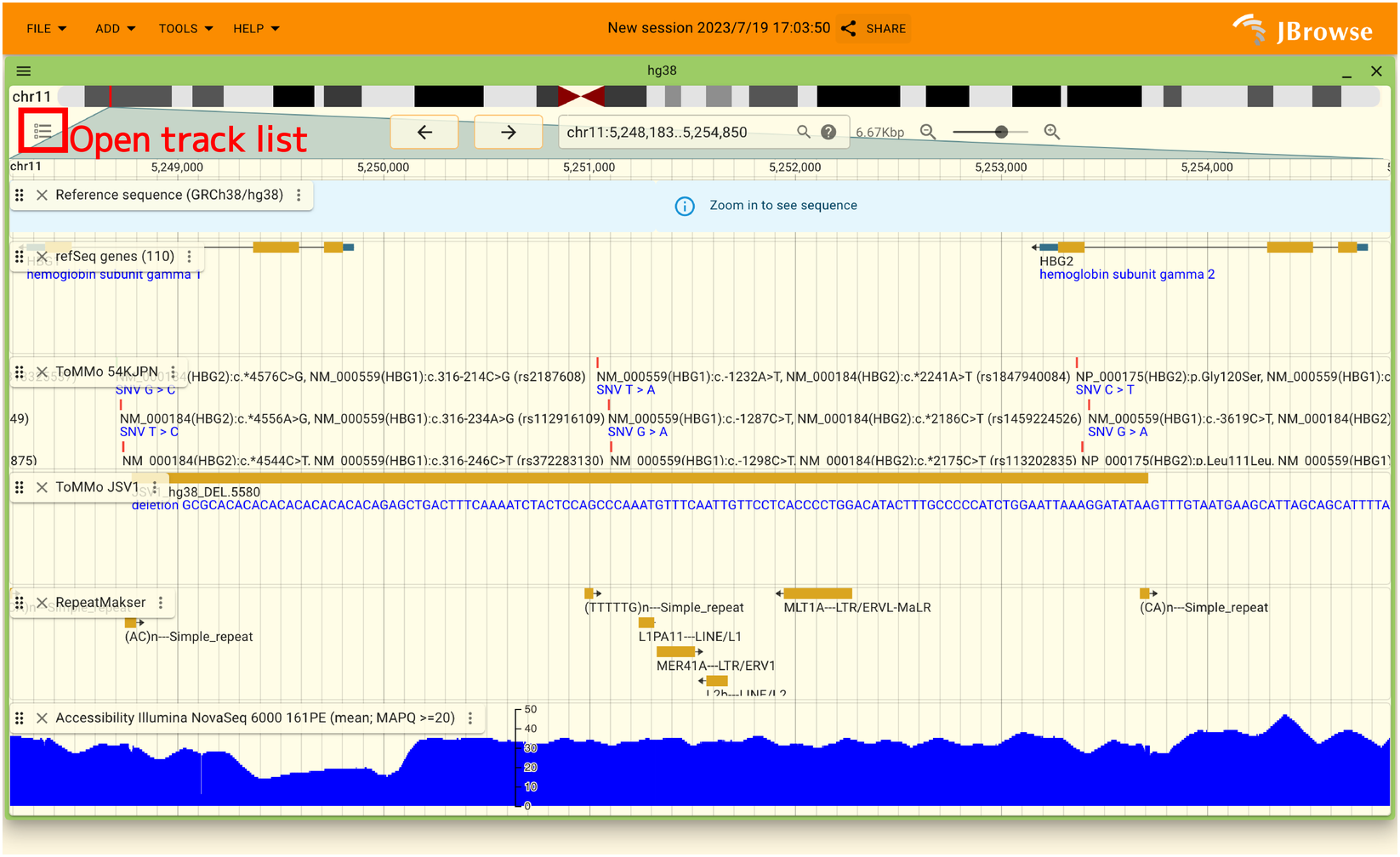
The list of tracks is opened as shown in the below.
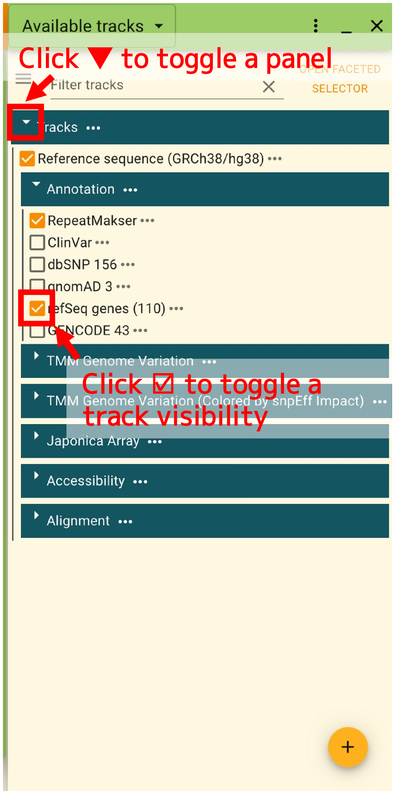
In this panel, you can select tracks to display. There tracks are classified by categories. The list of categories are shown in the below.
Category name |
Contents |
Assembly |
|---|---|---|
Annotation |
|
GRCh38/hg38, GRCh37/hg19, JG2.1.0 |
TMM Genome Variation |
|
GRCh38/hg38, GRCh37/hg19, JG2.1.0 |
TMM Genome Variation (Colored by snpEff Impact) |
|
GRCh38/hg38, GRCh37/hg19 |
Japonica Array |
|
GRCh38/hg38, GRCh37/hg19 |
Accessibility |
|
GRCh38/hg38, GRCh37/hg19 |
Variant co-occurrence |
|
GRCh38/hg38, GRCh37/hg19 |
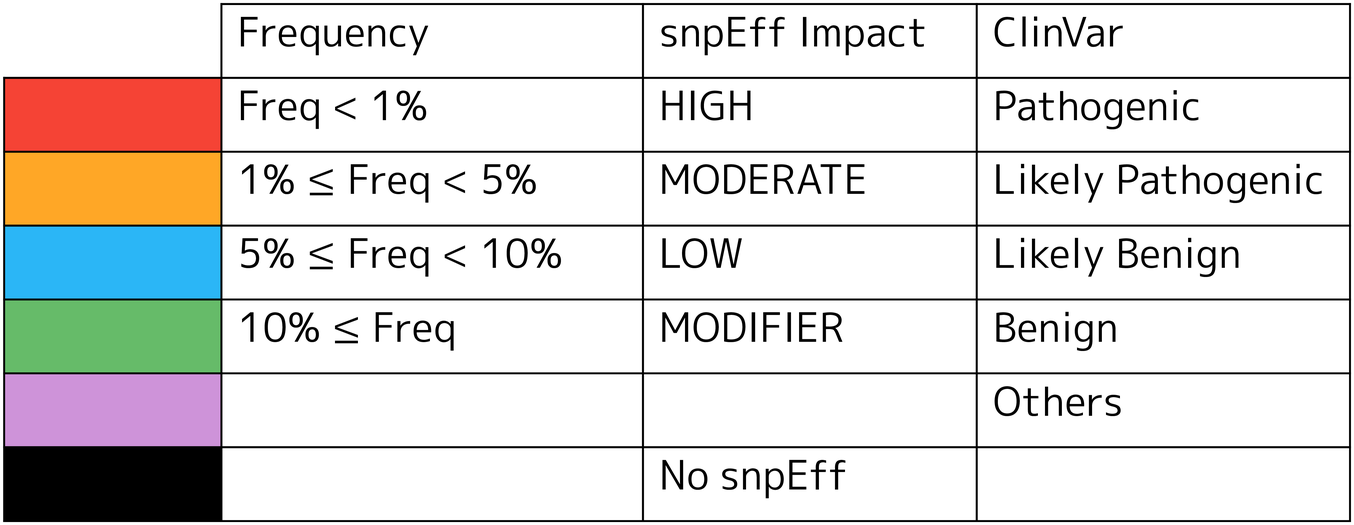
2.7.2.1. Color legend for variants
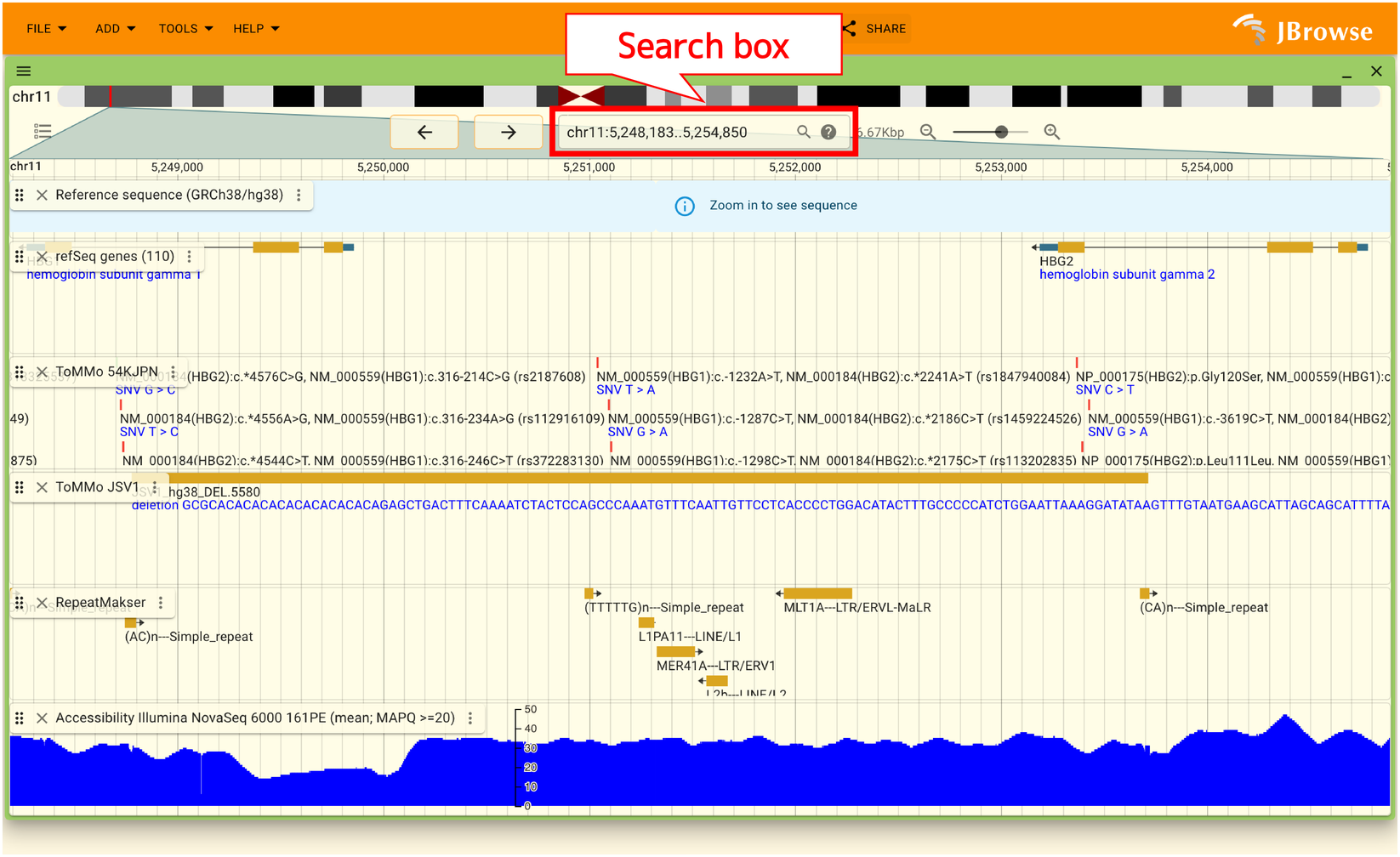
2.7.3. Region search
You can search a region by gene name, dbSNP ID, or amino acid change described with HGVS variant nomenclature. Searches can be performed from the box where the chromosome name and position are displayed, as shown in the image below.
Possible search items and examples are as follows
Search Item |
Example |
Note |
|---|---|---|
Gene symbol |
|
Searching by synonym (e.g. NRF2) is not supported. |
dbSNP ID |
rs671 |
|
amino acid change described with HGVS variant nomenclature |
|
|
When multiple areas are hit, the selection dialog will look like this.
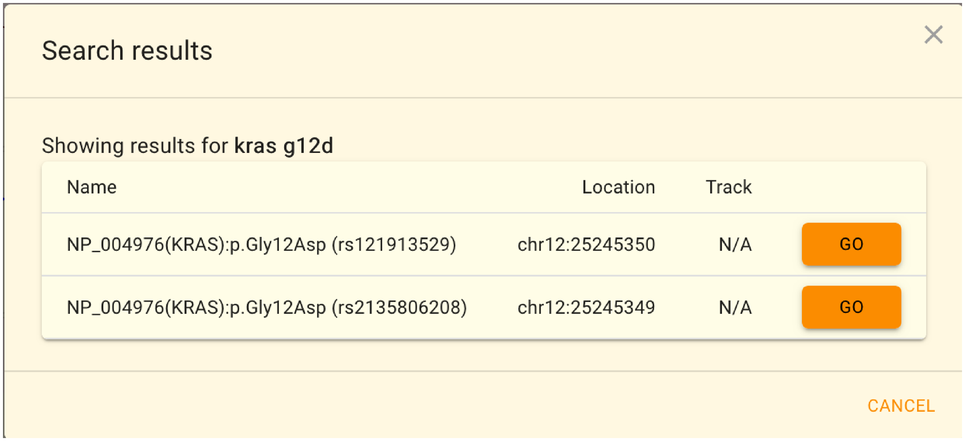
dbSNPのrs番号とHGVSによるアミノ酸変化での検索はGRCh37/hg19もしくはGRCh38/hg38でのみ利用可能であり、 jMorpのJBrowse2特有の機能となっています。JGシリーズでは遺伝子名の検索のみ利用可能です。
2.7.4. Overlay your data.
JBrowse2 allows you to overlay your own data on top of the data at your local machine. Your data will not be uploaded to the server, nor will Tohoku Medical Megabank Organization be notified of what data is being displayed.
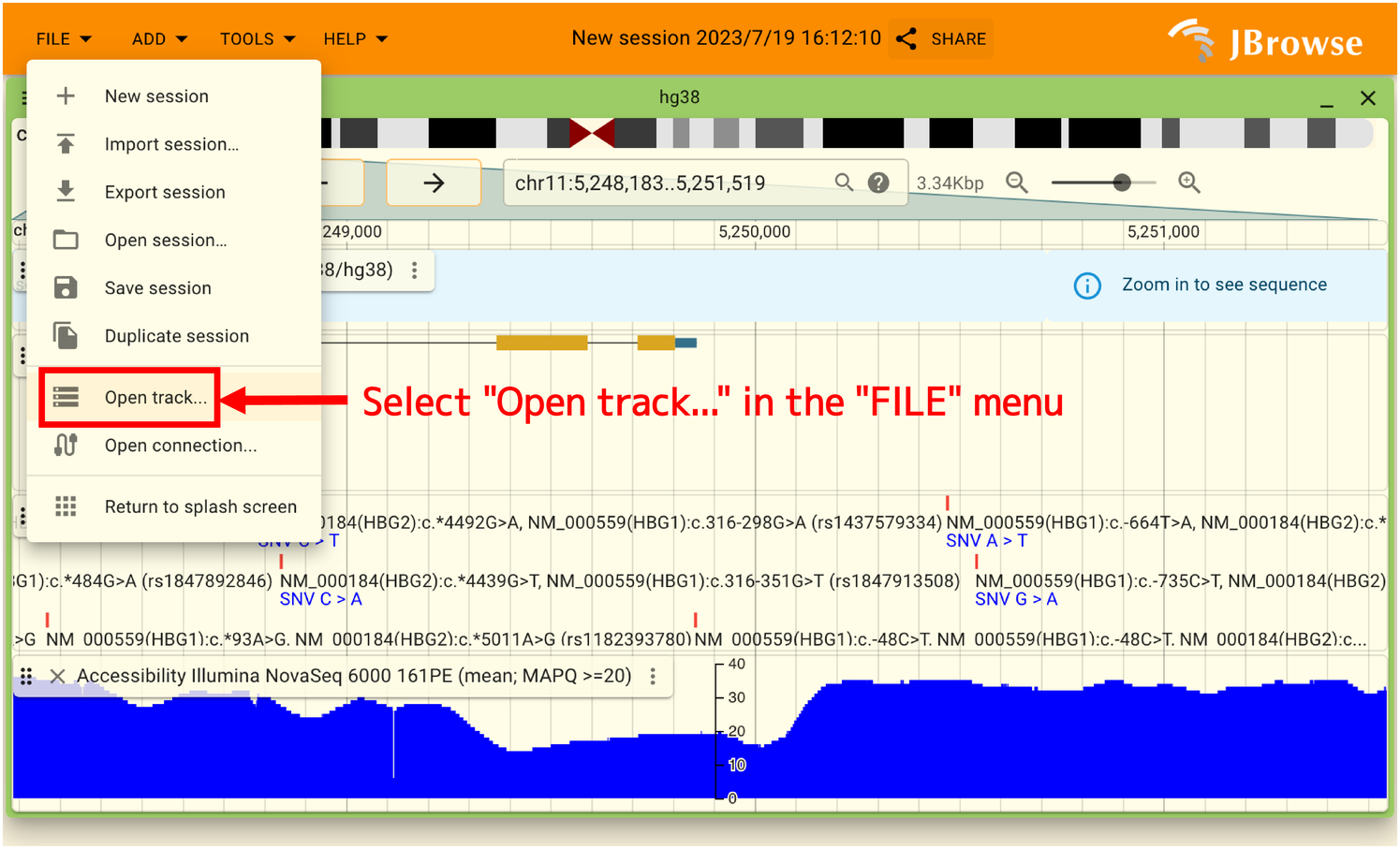
To overlay your data, select “Open track…” from the “FILE” menu from the “FILE” menu to display your data.
Next, enter the URL or select a file in the “Add a track” panel displayed on the right side. If the format requires an index file, it will not be loaded automatically like IGV, so you will need to select it manually. Click “NEXT” when done.
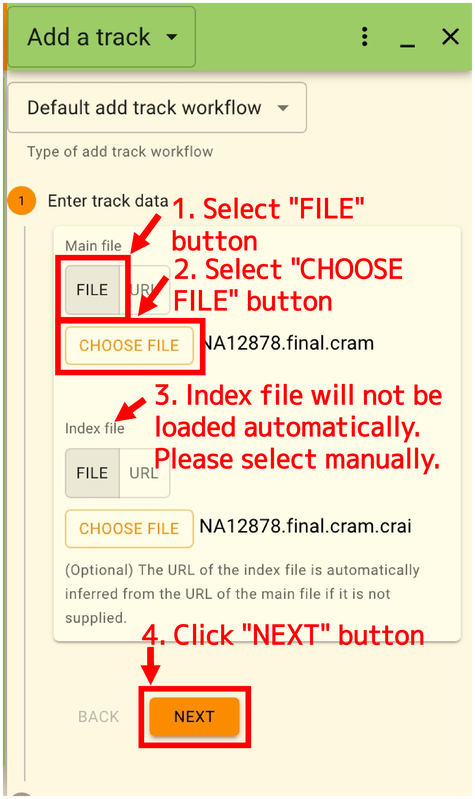
The settings are automatically suggested based on the contents of the file. If all is well, click “ADD”.
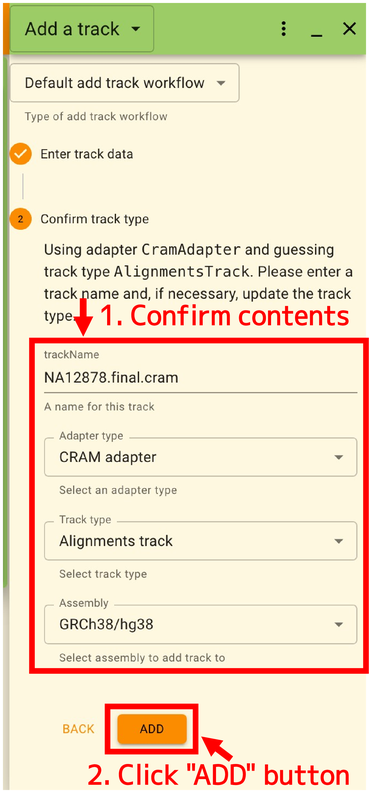
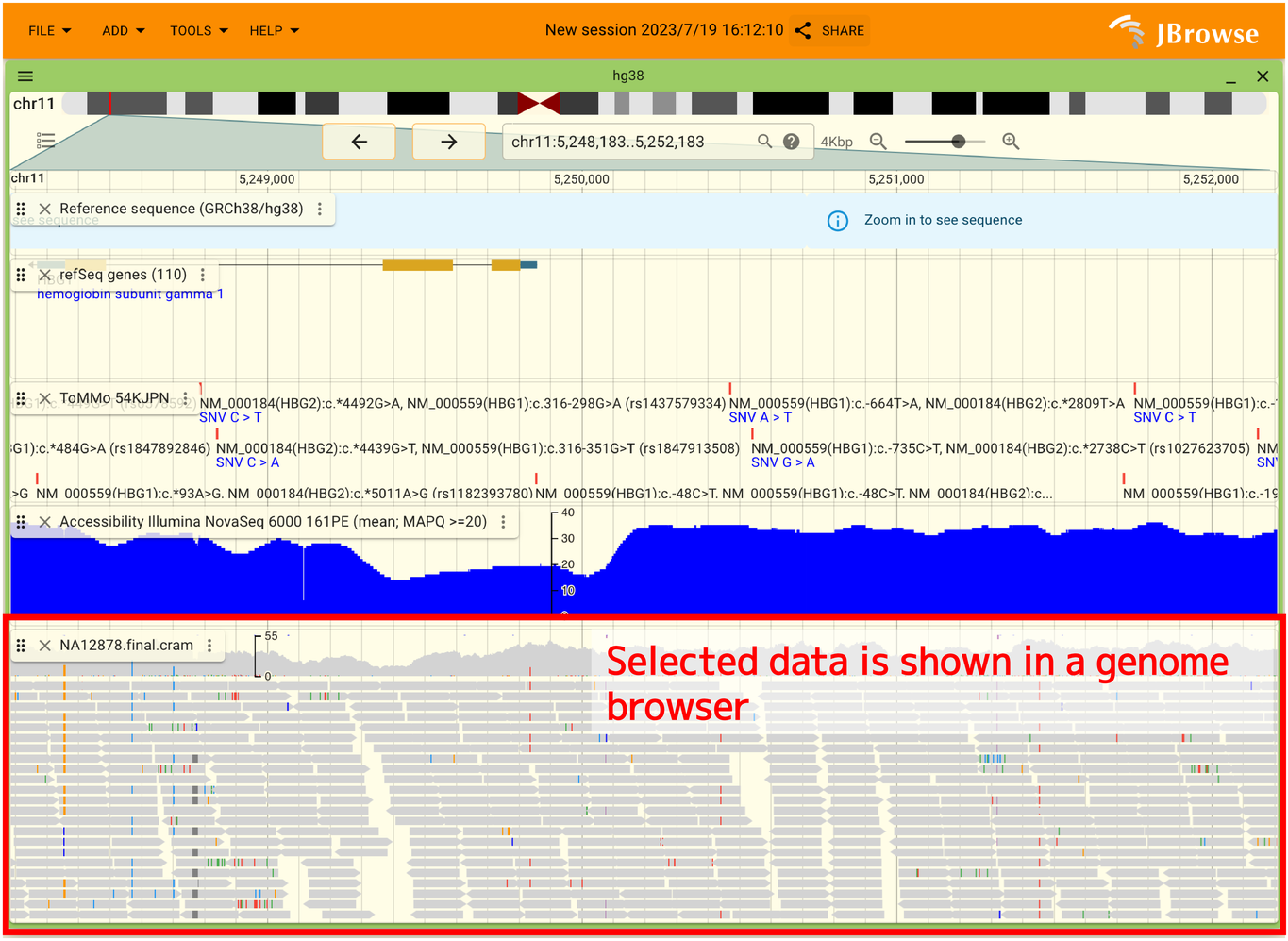
After performing the above operations, you can overlay the data at your local machine as shown below. In this example, a CRAM file is displayed, but BED and GFF files can also be displayed.
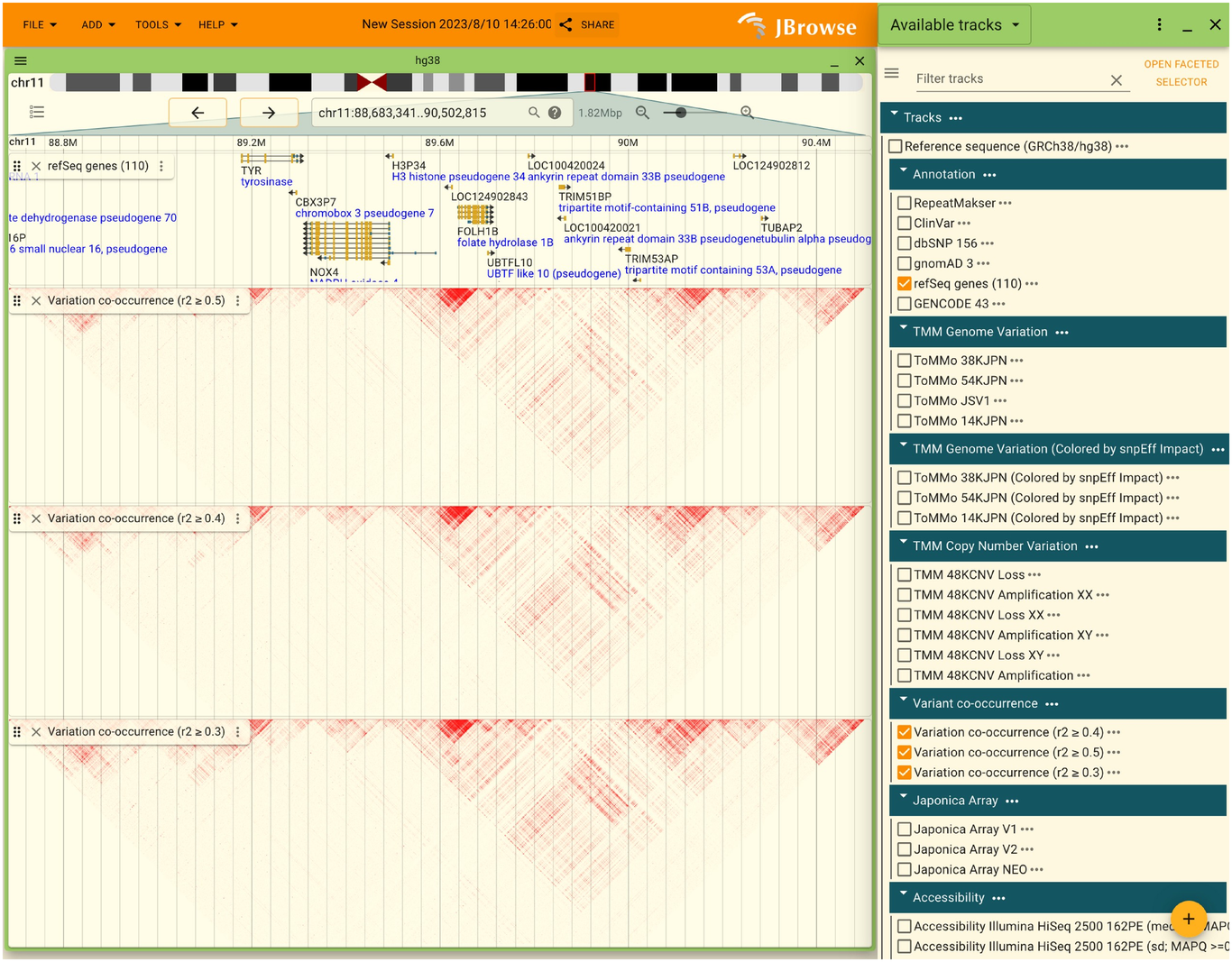
2.7.5. How to display variant co-occurrence
Variant co-occurrence can only be displayed correctly when a large enough area is displayed. If individual SNVs or INDELs are enlarged enough to be displayed, they should be reduced in size. If it is displayed correctly, it will look like this
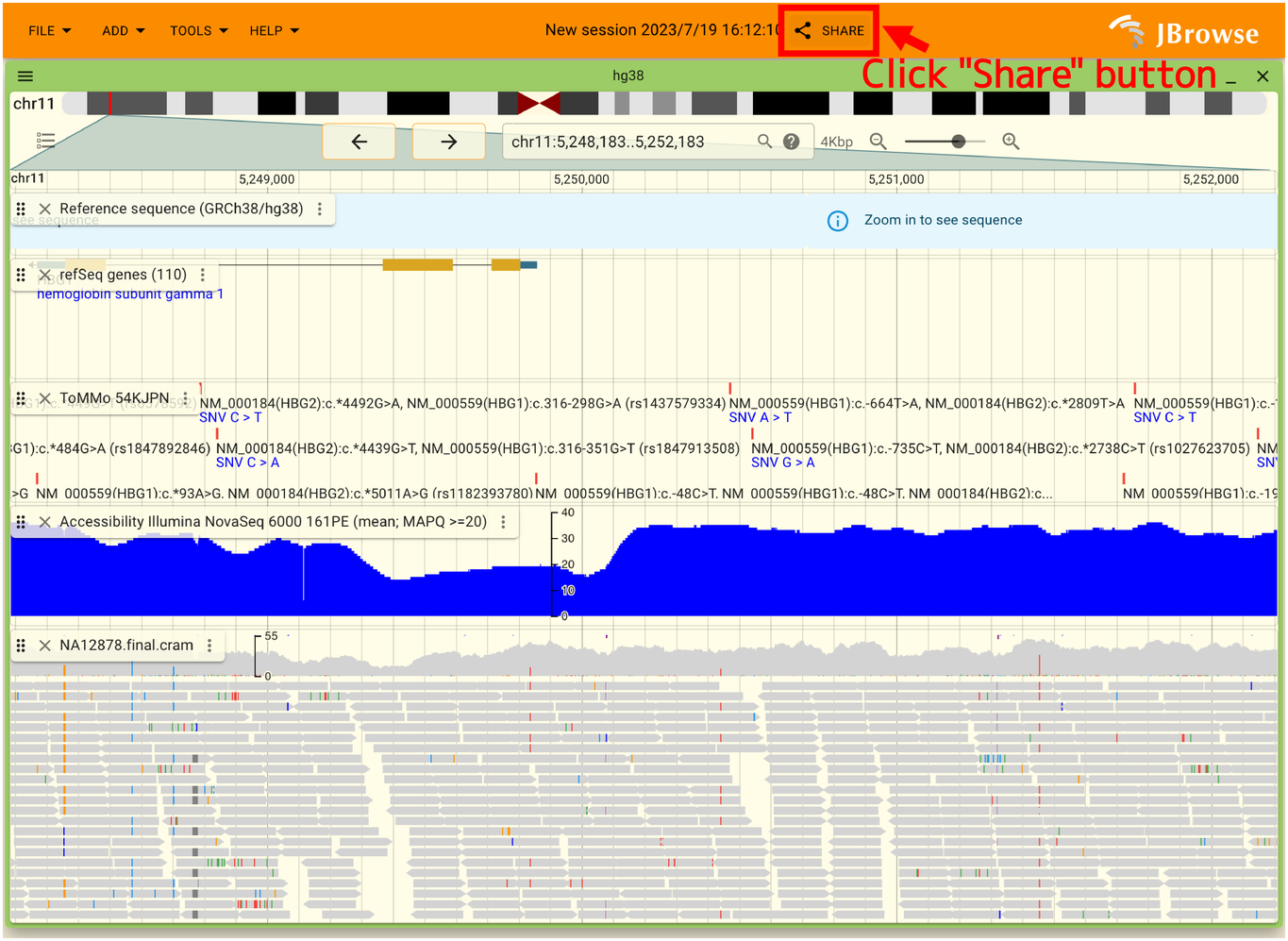
2.7.6. Share the contents of the display with others
JBrowse2 allows you to share the current display with others. When you do this, encrypted data of the current display is sent to the JBrowse developer. To share, click the “SHARE” button in the top header as shown below.
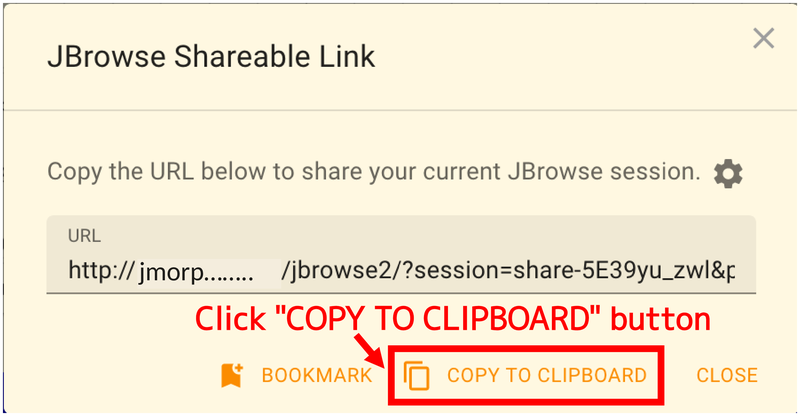
After a short wait when the dialog box appears, the URL for sharing will appear as shown below. Click “COPY TO CLIPBOARD” to copy the URL and share it.
2.7.7. Compare two reference genomes
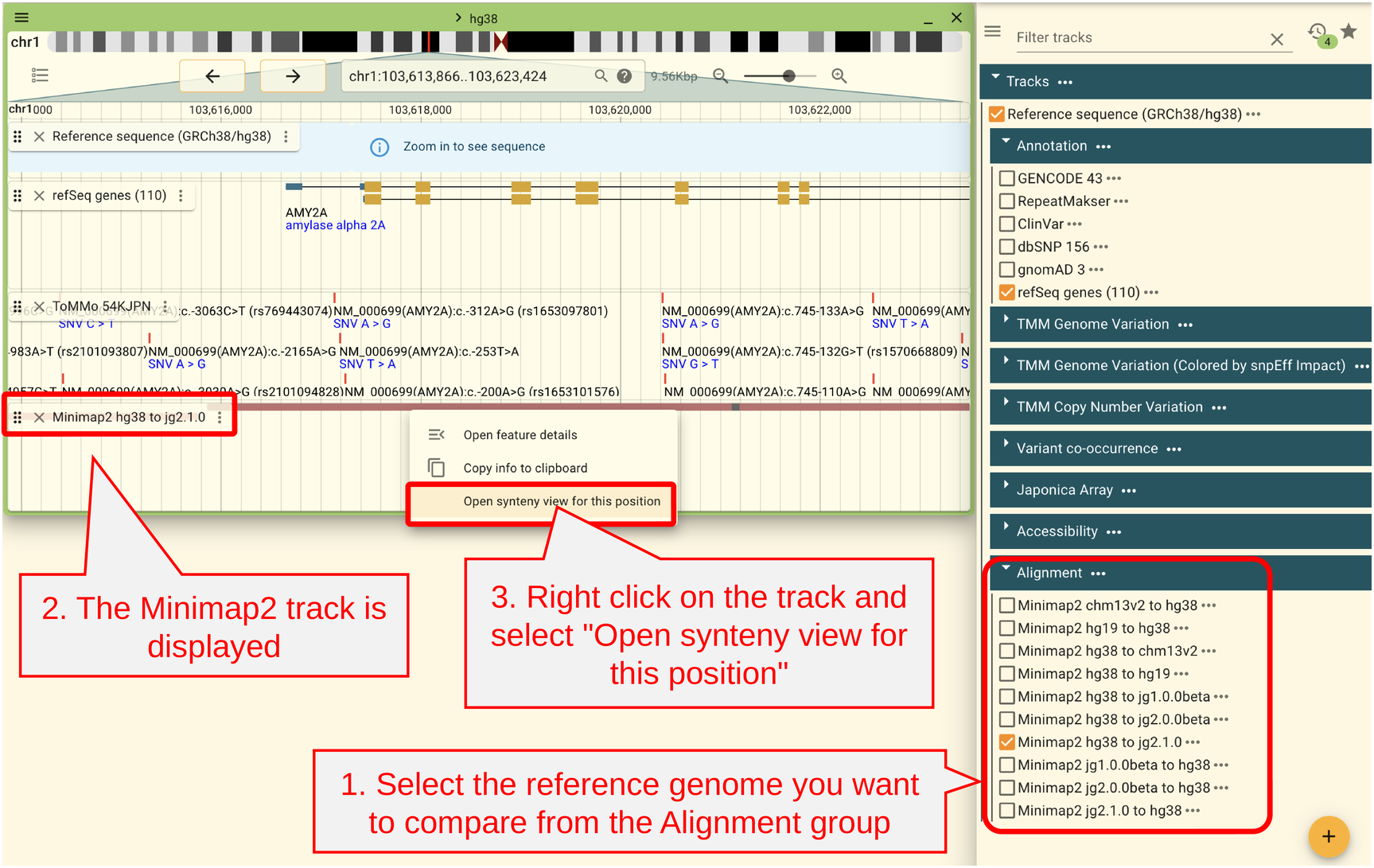
JBrowse2 allows comparison of two reference genomes.
First, open the track selector and open the track “Minimap2 XX to YY” in Aligment. XX is the reference genome currently open and YY is the reference genome to be compared. For example, if you have the hg38 reference genome open and want to compare it with JG2.1.0, select “Minimap2 hg38 to jg2.1.0”. Alignment using Minimpa2 is performed in two patterns, XX to YY and YY to XX, so both exist even for the same reference genome, and the results are slightly different. However, since the chain files for LiftOver are prepared for the XX to YY alignment, the XX to YY alignment should normally be used.
Right-click on the aligned region and select “Open synteny view for this position” from the menu.”
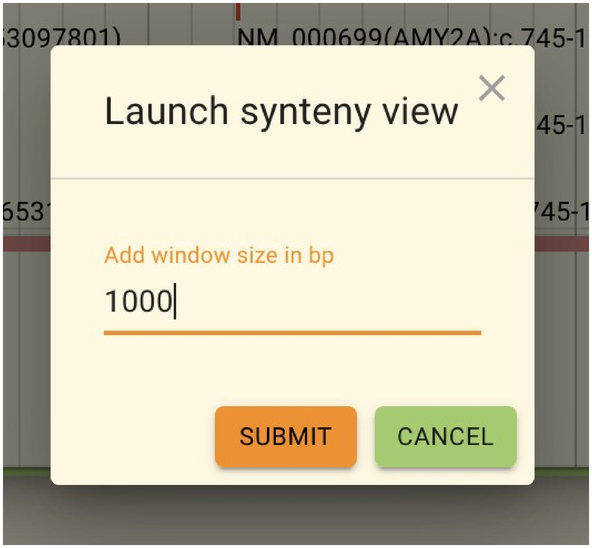
The following screen allows you to set how much to expand from the area you are currently viewing. If you are already viewing a large area, you can set the value to 0. After setting the values, click “Submit.”
In this state, no tracks are displayed for either reference genome. Therefore, please click “Open Track Selector” or select “View 1 track selector” or “View 2 track selector” from the menu outlined in red to open a track. Opening tracks such as refSeq or ToMMo Genome Variation will make it easier to compare the two reference genomes.
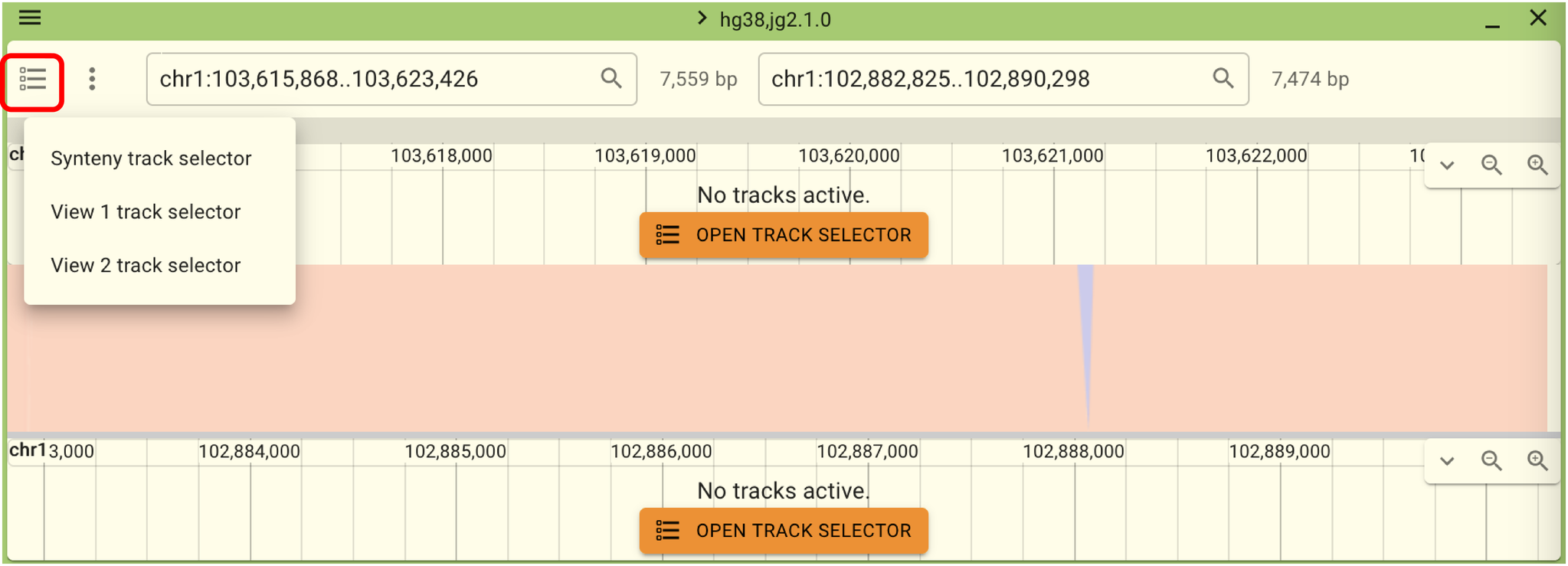
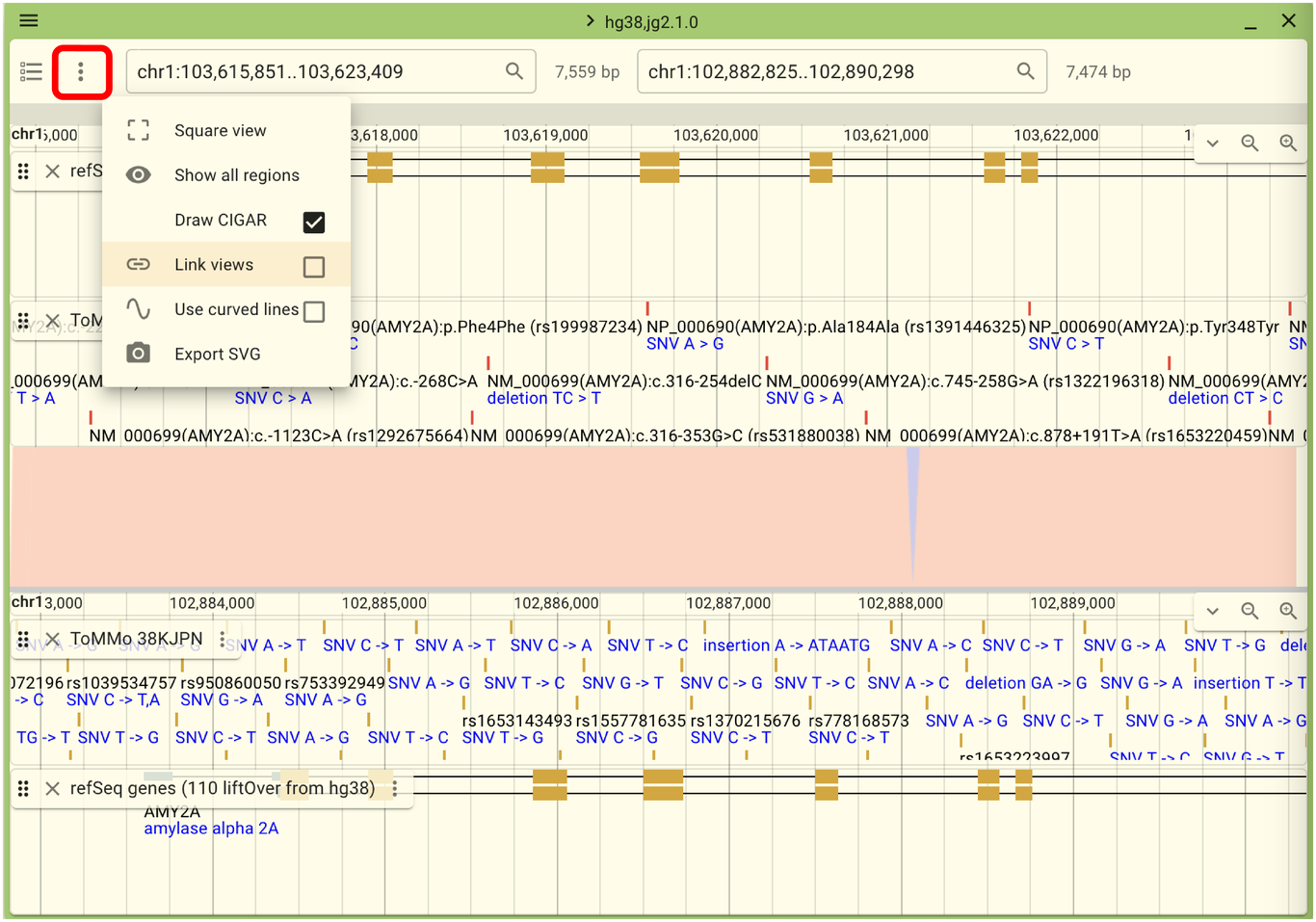
While in this state, moving the views left and right will not sync the two views. However, you can link the views by clicking the menu outlined in red in the following screen and selecting “Link views”.
2.7.8. JBrowse official document
Please refer JBrowse2 user guide for general use of JBrowse 2.