3.21. Metabolome 2022
- Dataset category
Metabolome
- Summary
Metabolome analysis results derived from approximately 53,000 Japanese individuals
- References
- Samples analyzed
Plasma
- Number of samples and analysis platform
Category
Analysis platform
Nunber of samples
NMR
NMR (Bruker: 600MHz),CryoProbe SampleJet54,956(Non pregnant: 40,529, Pregnant: 14,427, Repeat assessment: 4,599)LC-MS/G-Met v1
C18 column: UHPLC-Q-TOF/MS (Waters: Synapt G2-Si),HILIC column: HPLC-Q-FT/MS (Thermo Fisher Scientific: QExactive)1,266
LC-MS/G-met v2
HPLC-Q-FT/MS (Thermo Fisher Scientific: QExactive)
2,972
LC-MS/T-Met
UHPLC-MS/MS (Thermo Fisher Scientific: TSQ Quantiva)
2,365
LC-MS/kit180
UHPLC-MS/MS (Waters: Xevo TQ-S)
1,483 (Repeat assessment: 579)
LC-MS/kit500
UHPLC-MS/MS (Waters: Xevo TQ-S, TQ-XS)
9,231 (Repeat assessment: 1,338)
GC-MS/T-Met
GC-MS/MS (Shimadzu: TQ8040)
4,611 (Repeat assessment: 646)
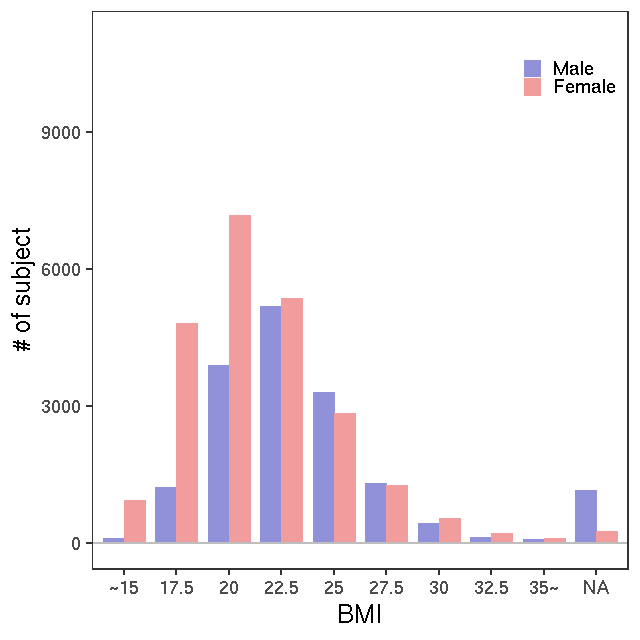
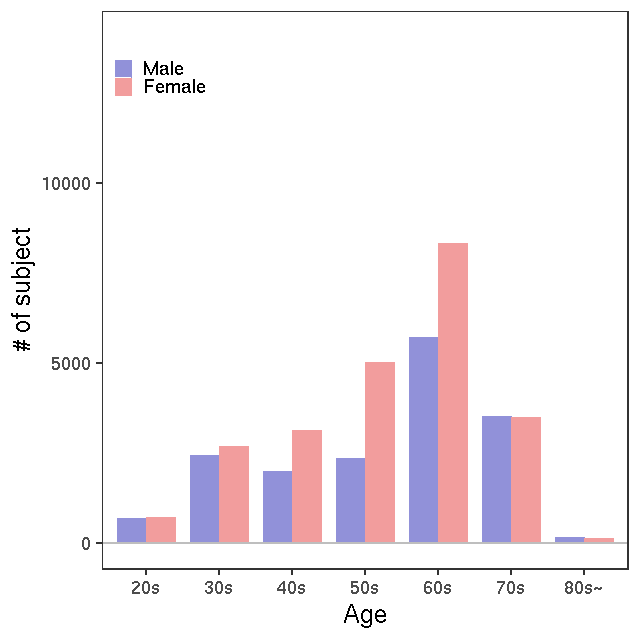
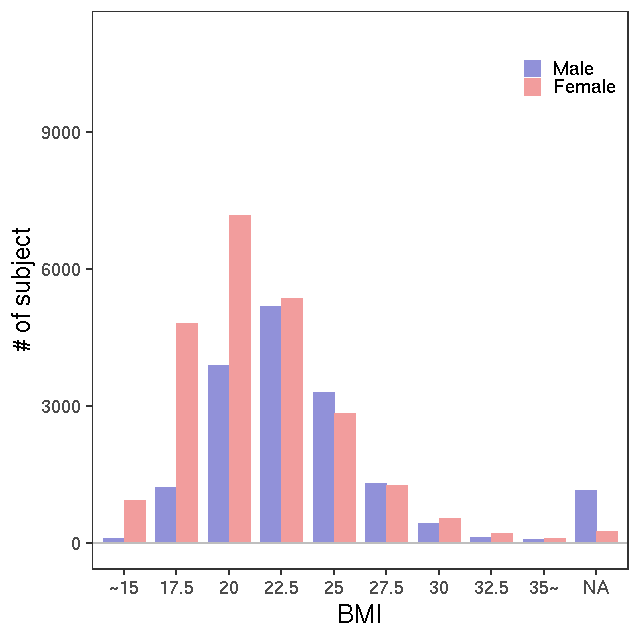
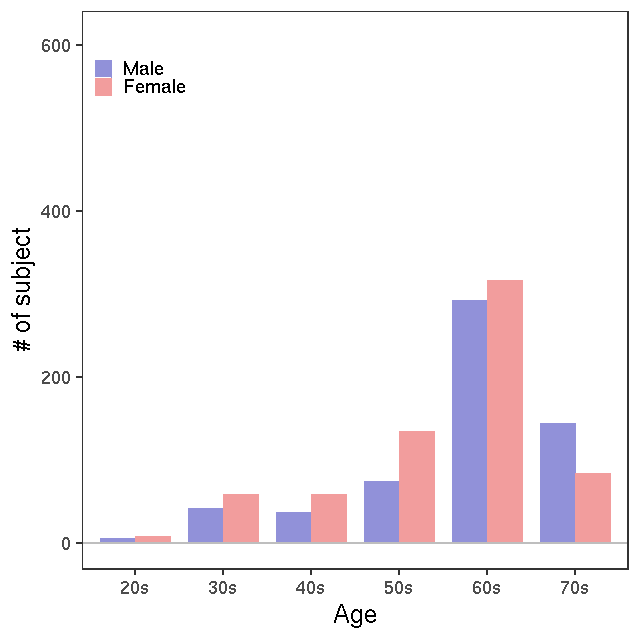
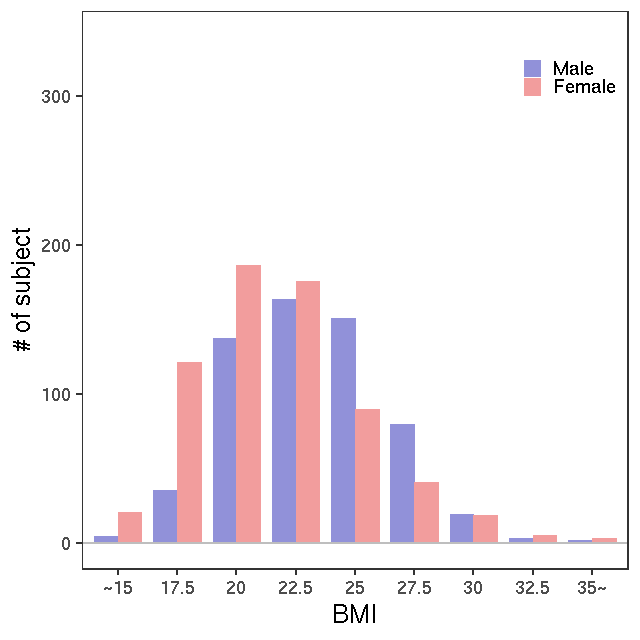
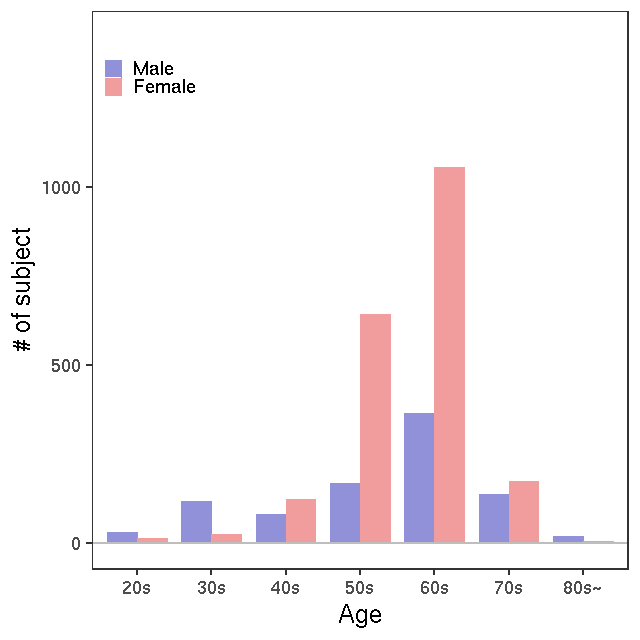
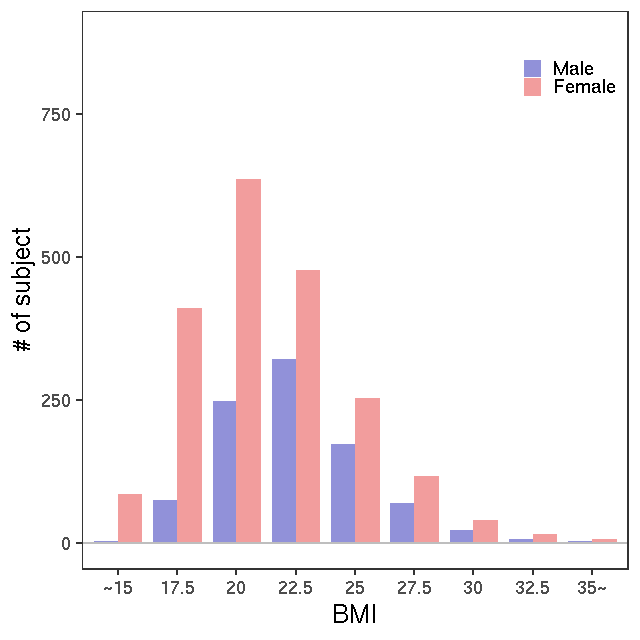
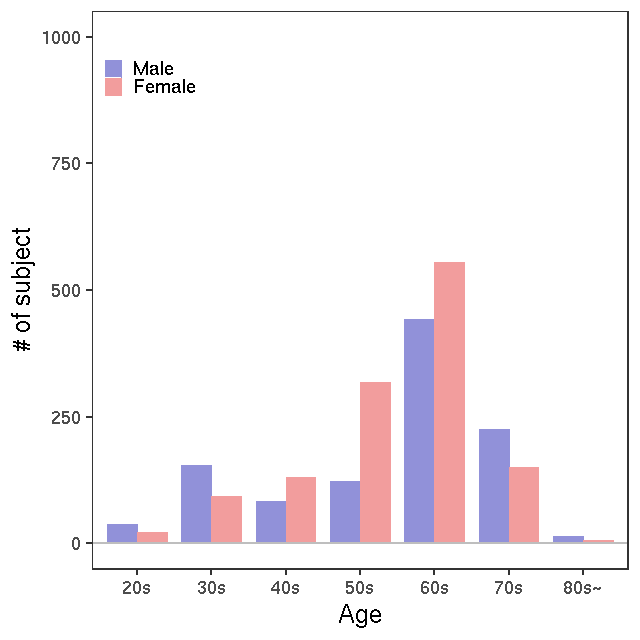
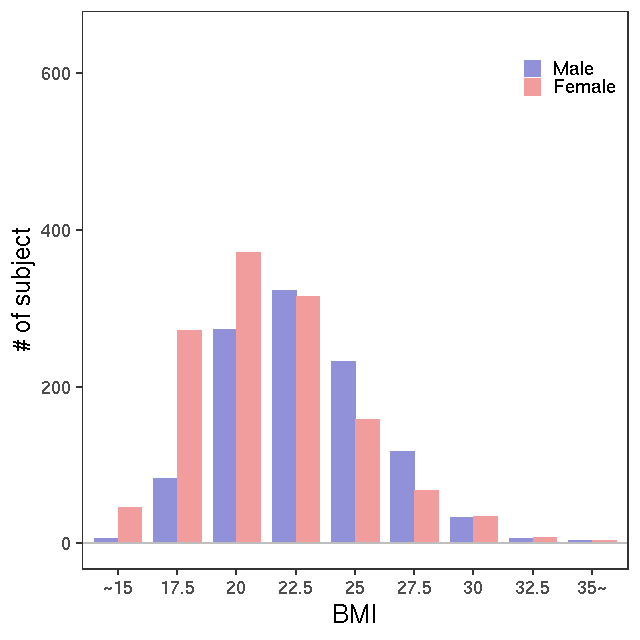
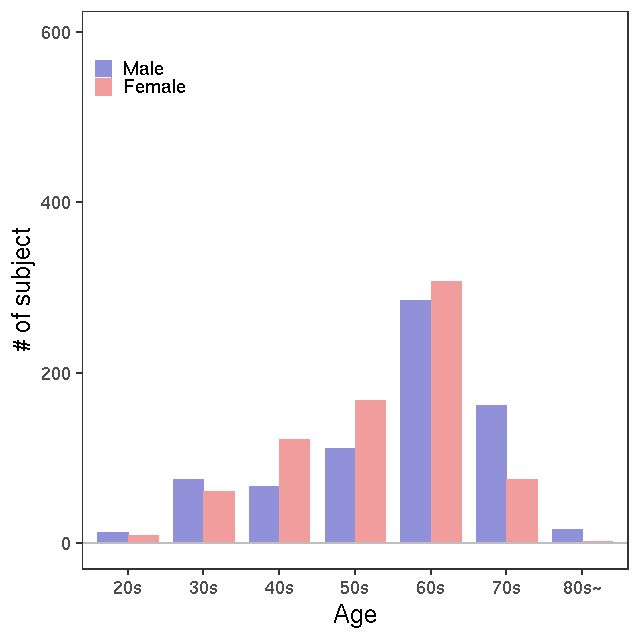
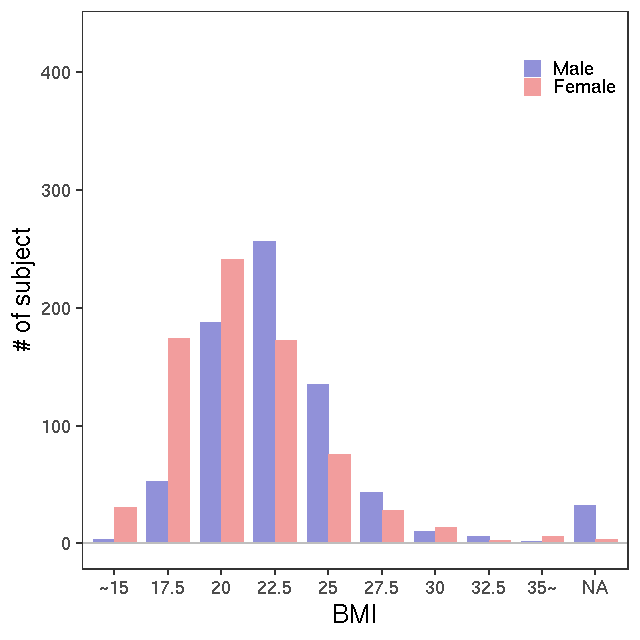
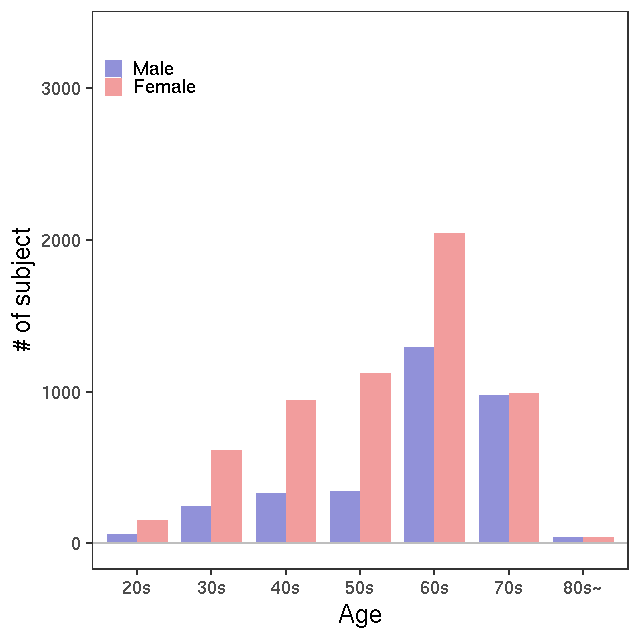
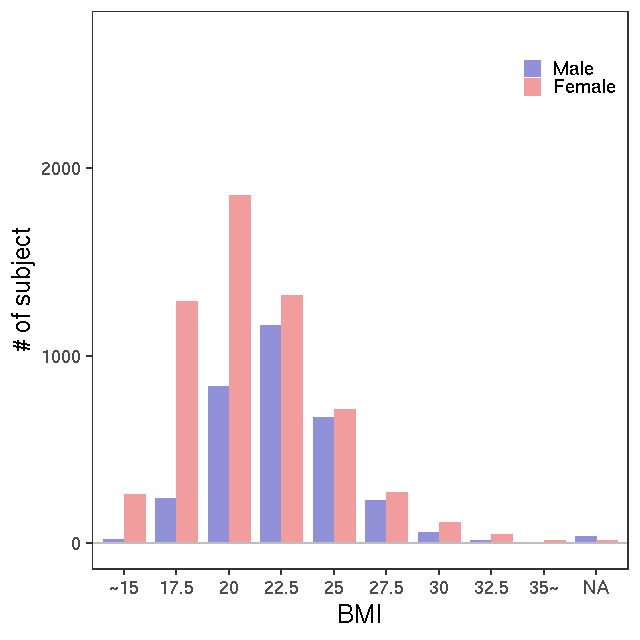
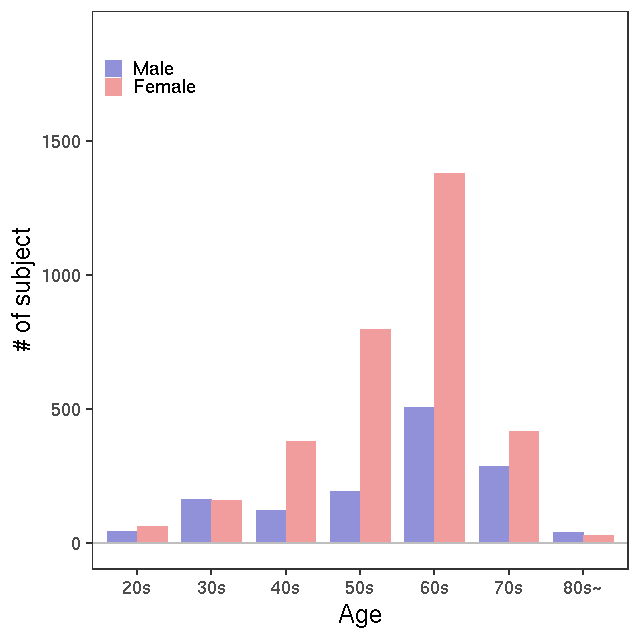
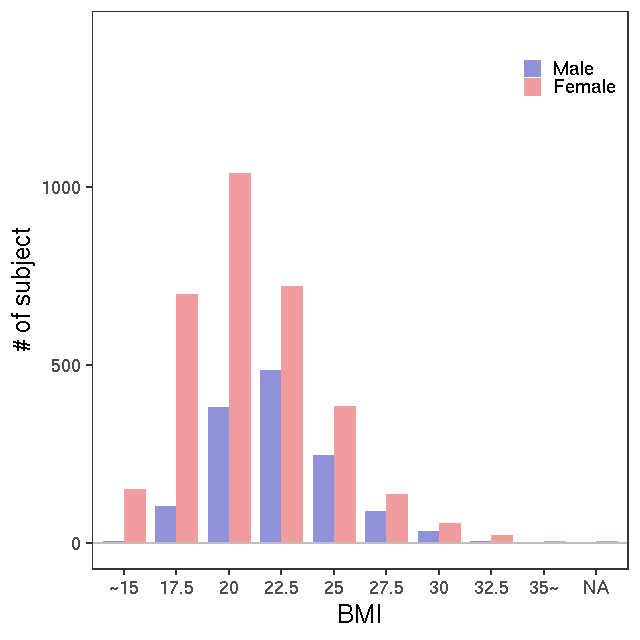
- Age / BMI distributions (non pregnant)
- Rule of metabolite ID convension
Metabolites in this dataset are assigned metabolite IDs in the form of TCx123456. The first two characters (TC) stands for ToMMo Compound ID (TC-ID). The third letter indicates how the metabolite is measured, and is divided as follows:
Prefix
Meaning
TCN
NMR
TCZ
LC-MS G-Met metabolome in HILIC mode ver.1 using HPLC
TCI
LC-MS G-Met metabolome in HILIC mode ver.2 using UHPLC
TCO
LC-MS G-Met metabolome in C18 mode using UHPLC
TCL
LC-MS T-Met metabolome
TCB
LC-MS T-Met metabolome in kit180
TCM
LC-MS T-Met metabolome in kit500
TCS
GC-MS T-Met metabolome
The last six digits are numbers assigned to each metabolite within each data source. In MS metabolome, data sources can be further divided into positive and negative modes. A number less than 500,000 indicates that the metabolite was detected in negative mode; otherwise, it is a metabolite detected in positive mode.
- Automatic quantification of metabolites
The concentrations of metabolites were automatically estimated from NMR spectra by using several regression models. Based on more than 1,000 concentration data carefully calculated by experts, both linear regression model and neural network model were built for each metabolite. We selected the model with best performance by using R-squared (R2) values as an evaluation index. We provide a reliability score of estimated concentration on a four-tiered scale: “Triple Stars (★★★)”, “Double Stars (★★☆)”, “Single Star (★☆☆)” and “Zero Star (☆☆☆)”. Each category corresponds to R2 value >=0.9, >=0.7, >=0.6, and <0.6, respectively. Outliers were defined by this protocol as those concentration is >10 SD after automatically estimation and were excluded in each compound.
- Note related to Hypoxanthine and Inosine
Because values of metabolites, (TCN000044, TCI006689, TCS000091, TCO501589, TCZ000947) and Inosine (TCN000045, TCI010703), strongly depend on the stored time until specimen processing for banking, values of the samples that were not processed on the day were excluded for these two metabolites.
- Related pages on jMorp website