2.13. GWAS page
jMorp has a repository for storing the results of GWAS analyses performed within the TMM project. This repository contains summaries of the GWAS studies (publication, study overview, analysis method) and summary statistics files. The summary statistics files that can be downloaded from the jMorp GWAS pages can be used for meta-analysis.
2.13.1. Structure of GWAS pages
The GWAS layer in the jMorp consists of the following subpages
Page name |
Page contents |
|---|---|
GWAS study list |
|
GWAS study details |
|
GWAS trait list |
|
GWAS trait details |
|
Details of GWAS analysis results |
|
Each page is linked as follows.
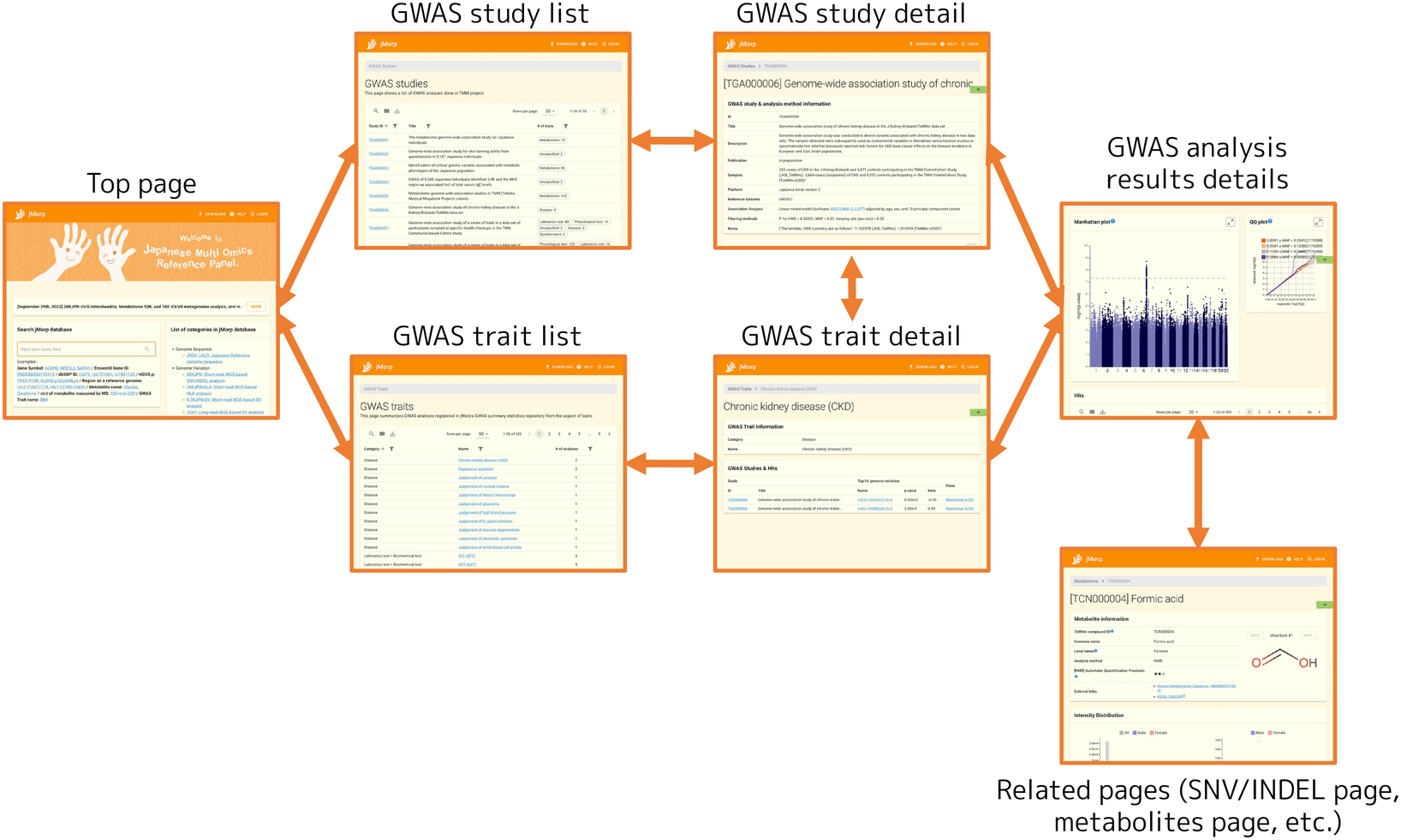
Following the GWAS studies and GWAS traits links at the bottom right of the jMorp top page will take you to the GWAS study list page and the GWAS trait list page, respectively.
2.13.2. GWAS study detail page
The GWAS study detail page displays a summary of the GWAS study and a list of GWAS analysis results included in the study. Here, the contents of a GWAS study detail page will be explained by using TGA000001 (https://jmorp.megabank.tohoku.ac.jp/gwas-studies/TGA000001) as an example.
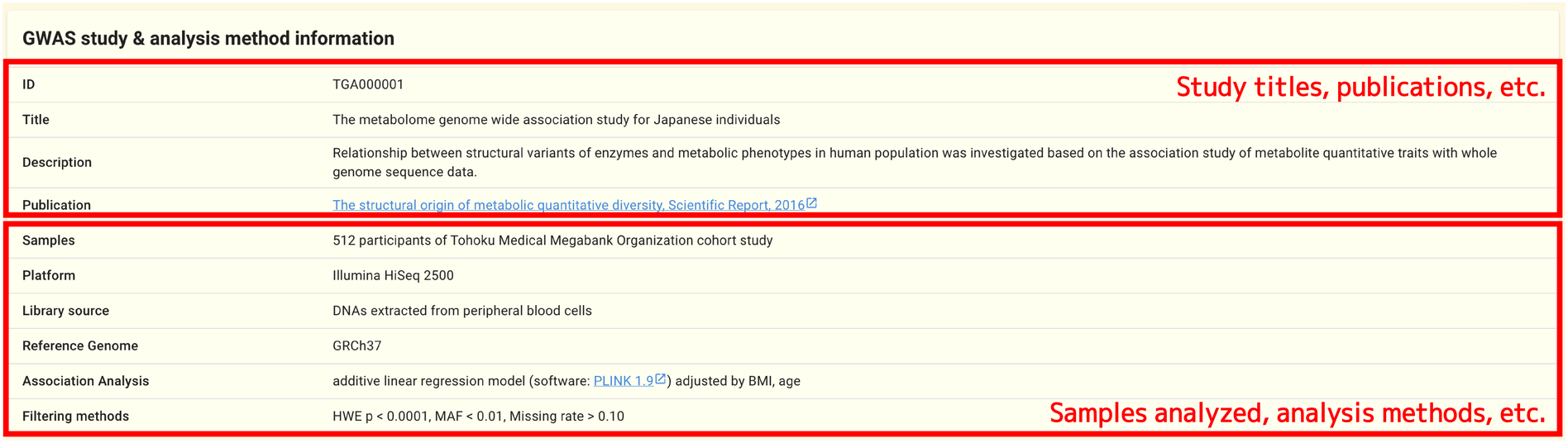
An summary of the GWAS study and analysis methods can be found in the information panel at the top of the page.
You can find a table of the analysis results included in this study at the bottom of the page.
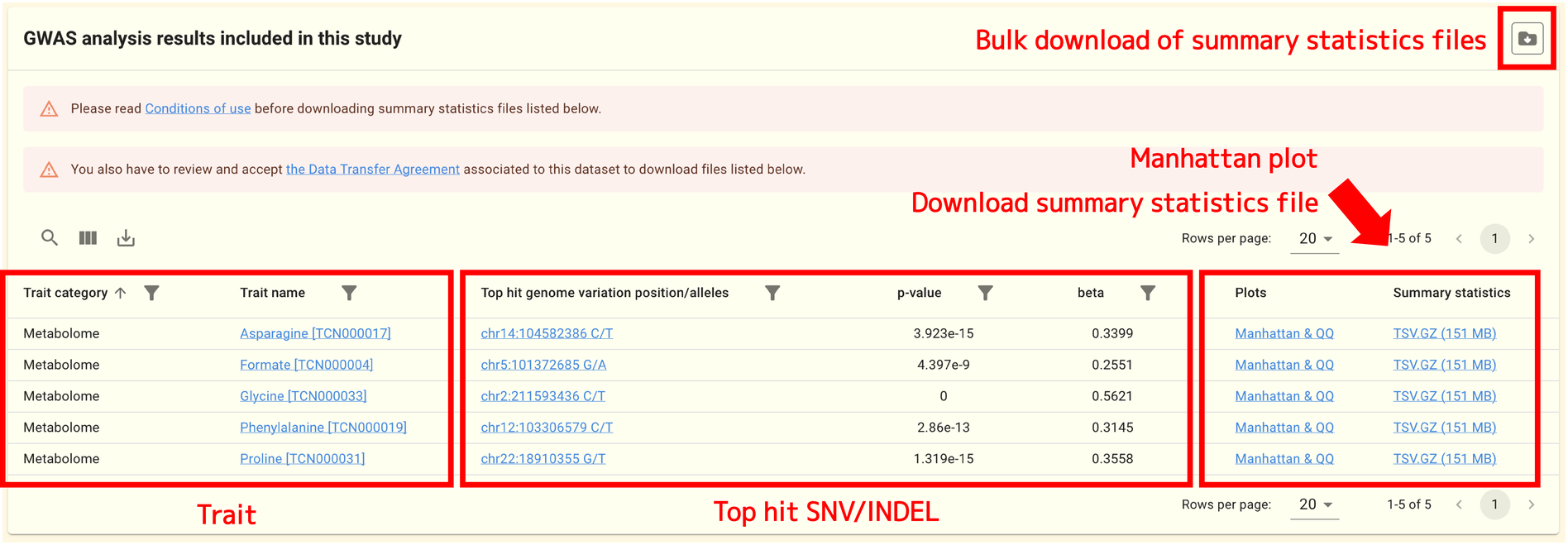
In principle, one trait corresponds to one row of the table. The trait name, top-hit SNV/INDEL, p-value, and beta value are shown for each trait. You can visit the SNV/INDEL page to view the details of this SNV/INDEL by clicking the SNV/INDEL link.
The analysis detail page, where you can view the Manhattan plot and other information, can be accessed by clicking the Manhattan & QQ link in the table. By clicking the TSV.GZ link, a file containing summary statistics can also be downloaded.
Note
You must read and accept the Data Transfer Agreement (DTA) linked to each GWAS study in order to download the summary statistics files with summary statistics.
2.13.3. GWAS analysis result detail page
You can view the summary statistics file derived from the GWAS analysis on the GWAS analysis results page.
The top of the page shows the Manhattan plot and QQ plot generated from the summary statistics file. Clicking the buttons in the top right corner of each panel will enlarge the plots.
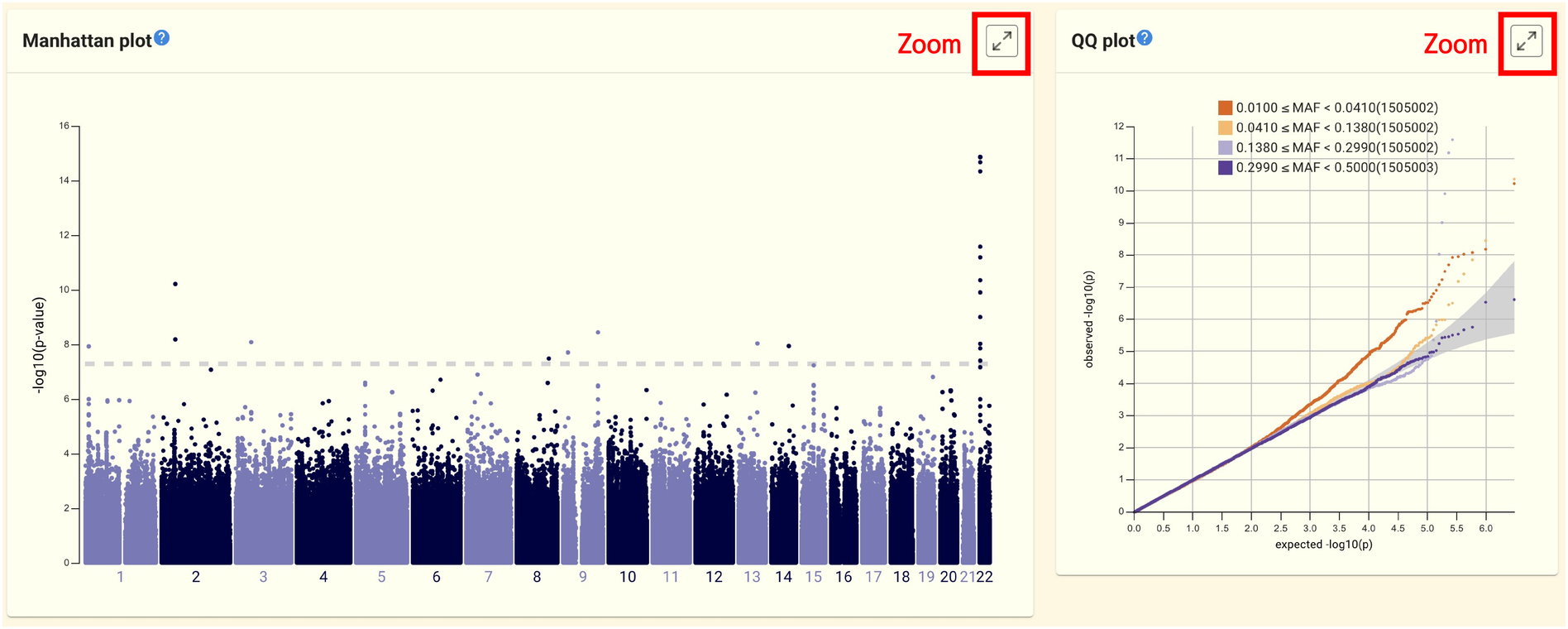
A table of SNV/INDELs that were significant over the threshold can be found at the bottom of the page. From here, you can go to jMorp’s SNV/INDEL page to see more about SNV/INDEL in-depth.
2.13.4. GWAS trait detail page
jMorp’s GWAS repository manages data for each GWAS study. Multiple GWAS studies may analyze the same trait (with different target populations and analysis methods). The GWAS trait detail page allows you to list the results of GWAS analysis done for the same trait.
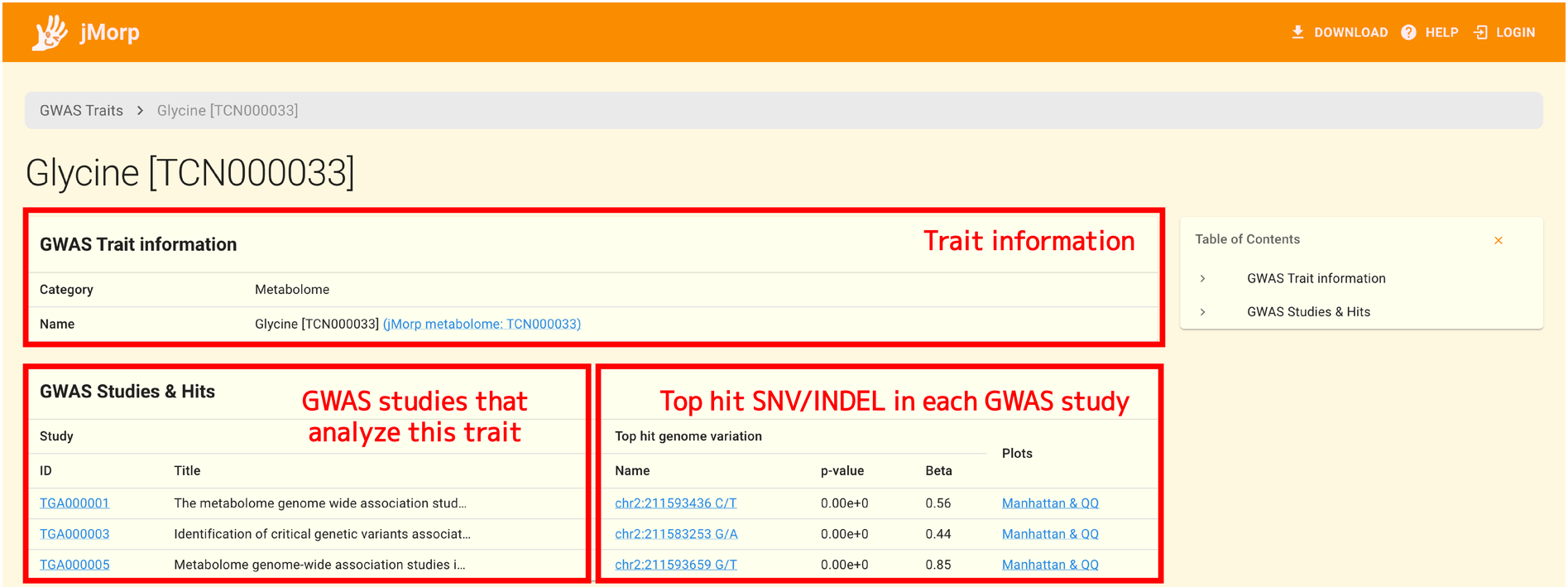
The trait name and links to relevant entries in jMorp are displayed in a panel at the top of the page.
The list of GWAS studies that analyzed this trait is located at the bottom of the page, along with details on the SNV/INDEL that was identified as the top hit in each of those analyses.