2.4. SNV/INDEL protein structure mapping tool
2.4.1. Basic usage
jMorp contains protein structure mapping data that shows the location of genome variation (SNV/INDEL) in protein structures. This is done by sequentially converting SNV/INDEL from the position on the DNA sequence to the position on the RNA sequence and then the position on the amino acid sequence, and then using the obtained sequences to generate the Protein Data Bank (PDB) and AlphaFold DB by executing a BLAST search against the amino acid sequence database to specify the position on the protein.If mapping data exists, the SNV/INDEL protein structure mapping tool can be accessed from the gene page or SNV/INDEL page to see the positional relationship between SNV/INDELs and protein structures.
The figure below shows the structure mapping data for rs671 onto PDB protein structures .
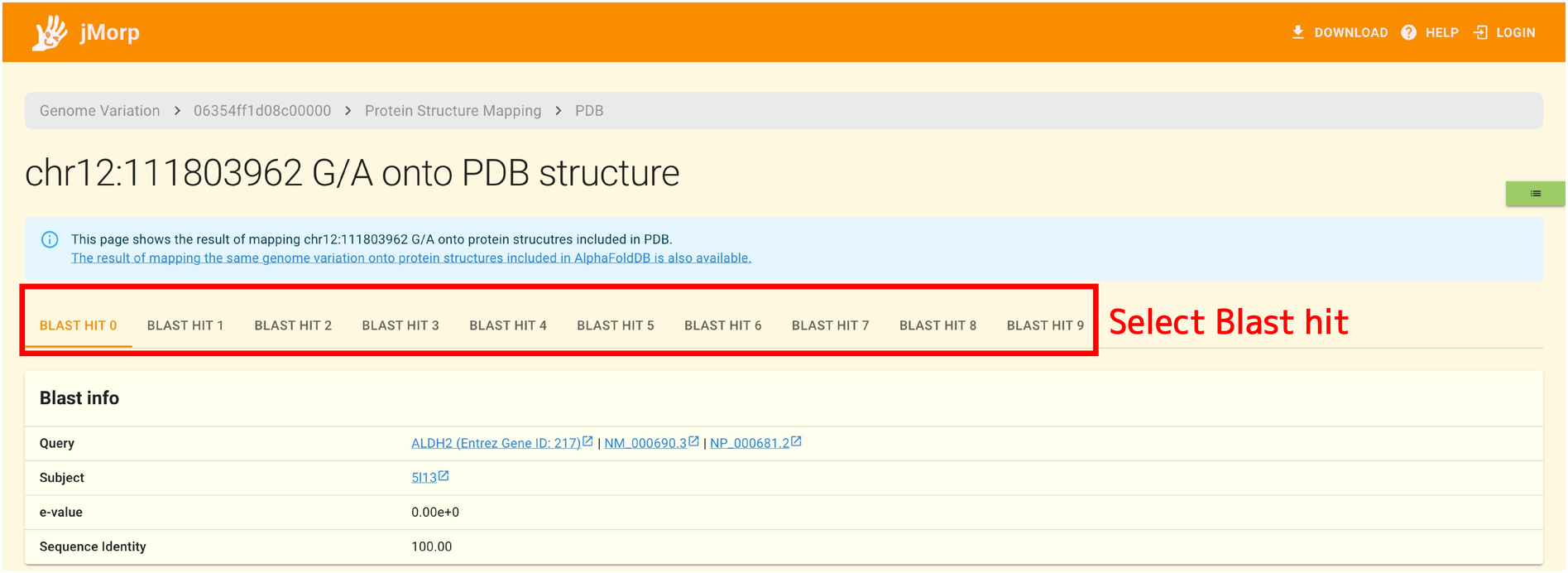
At the top of the page, there are tabs labeled BLAST HIT 0, BLAST HIT 1 … As mentioned above, the final step is to map SNVs and INDELs to protein structures by determining the similarity between sequences by Blast. In this Blast process, one amino acid sequence may be matched to multiple protein structures. You can switch between matches by using the BLAST HIT tab at the top of the page.
The Blast Info panel under the tab displays the results of the aforementioned Blast search (a sequence given as a query, protein structure sequences matched, e-values, sequence identities, etc.).
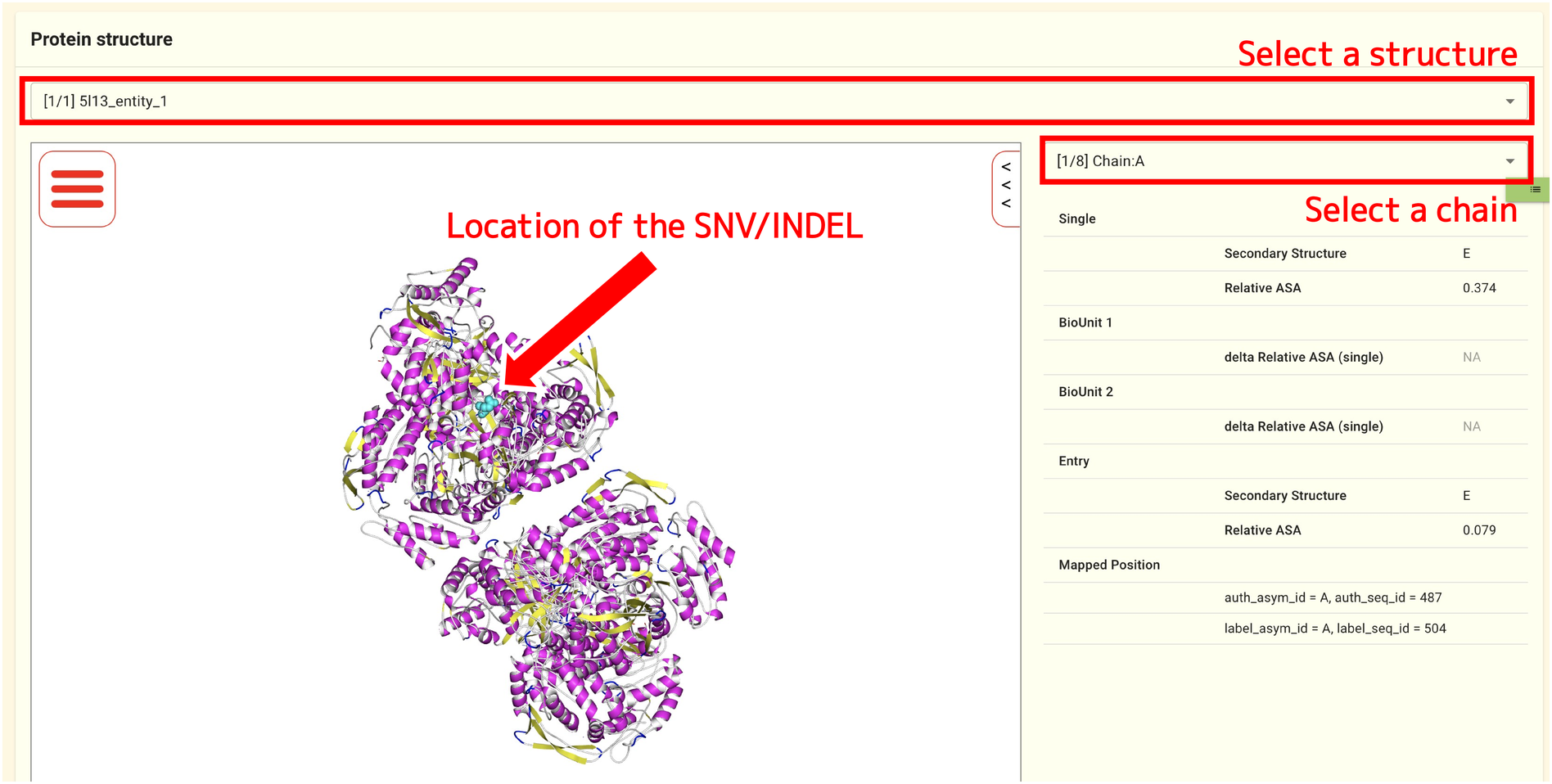
You can see the location of the SNV/INDEL by looking at the protein structure model in the panel at the bottom of the page. You can select a protein structure using the selector at the top of the panel. The jMorp protein structure viewer groups protein structures in the PDB by amino acid sequence similarity, so that protein structures with the same amino acid sequence appear as a single Blast Hit. The selector at the top of the Protein structure allows you to choose which of the grouped protein structures to display the SNV/INDEL mapping results for.
The right side of the Protein structure panel displays a selector to select a chain, the properties of the structure (secondary structure, solvent-exposed surface area (ASA), etc.), etc.
The results of the SNV/INDEL mapping for the chosen protein structure or chain are shown on the left side of the Protein structure panel. On the protein structure shown in the ribbon model, the locations of SNVs and INDELs are shown as light blue spheres. By using the mouse, you can rotate, enlarge, or shrink the protein structure.
2.4.2. Switch to the mapping results page for protein structures in AlphaFold DB
jMorp contains the results of mapping SNV/INDEL to protein structures in PDB as well as the results of mapping SNV/INDEL to proteins in AlphaFold DB.
You can get the results of mapping to proteins in the AlphaFold DB by using the switching link that is displayed at the top of the page if the data are available.
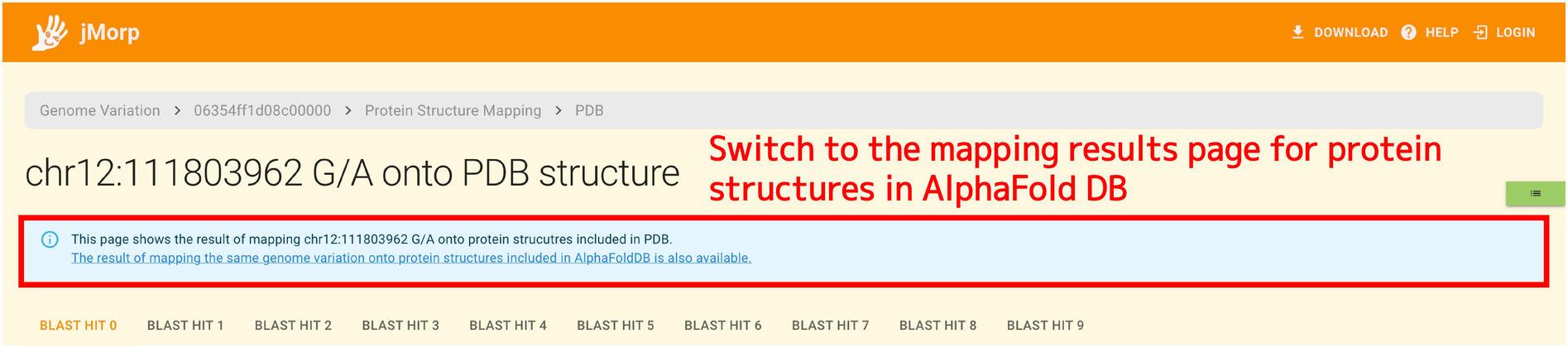
The content that is displayed as the mapping result is equivalent to the PDB mapping result.