2.16. Tips: Preference page
To access the Preferences page, choose PREFERENCE at the top of the jMorp website. You can alter the jMorp website’s appearance according to your preference using the settings on this page. There are three items that can be adjusted as of this writing:
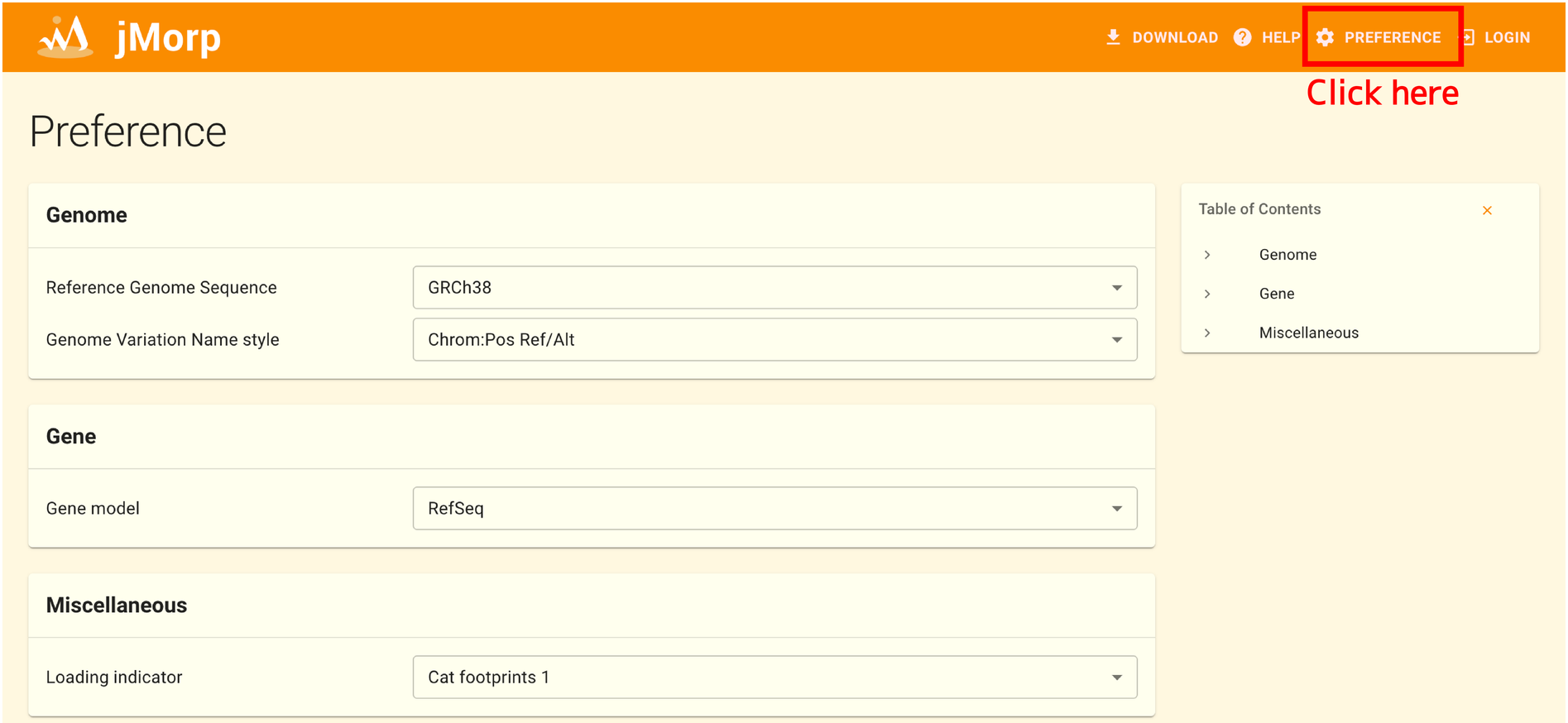
2.16.1. Genome → Reference Genome Sequence
A preferred reference genome sequence can be selected. You have a choice between GRCh37 and GRCh38; GRCh38 is the default.
For example, if this item is set to his GRCh38, searching for rs671 in the jMorp search box will result in chr12: 111,803,962 G/A (GRCh38), and chr12: 112,241,766 G/A (GRCh37) is not shown in the search results. In addition, if both GRCh37 / GRCh38 data exist for the same data set, the data employs the coordinate system you choose in this option will be shown.
2.16.2. Genome → Genome Variation Name style
Select how to express SNV/INDEL. You can choose from Chrom:Pos Ref/Alt or HGVS, default is Chrom:Pos Ref/Alt.
When this item is set to Chrom:Pos Ref/Alt, for example rs671 will be displayed as chr12:111,803,962 G/A (GRCh38). On the other hand, when this item is set to HGVS, rs671 is represented as NP_000681(ALDH2):p.Glu504Lys. The HGVS notation is based on the Refseq canonical transcripts. For SNVs/INDELs not in the gene region are notated in HGVS.g style like chr12:g.111803962G>A.
2.16.3. Gene → Gene model
Select the gene model to be used in the gene annotation panel on the SNV/INDEL page. You can choose from RefSeq or GENCODE, default is RefSeq.
2.16.4. Miscellaneous → Loading indicator
You can select an animation to be shown before the page has finished rendering.